Evolution of Fungal U3 snoRNAs: Structural Variation and Introns
Abstract
:1. Introduction
2. Methods
2.1. Homology Search
2.2. Secondary Structure Prediction
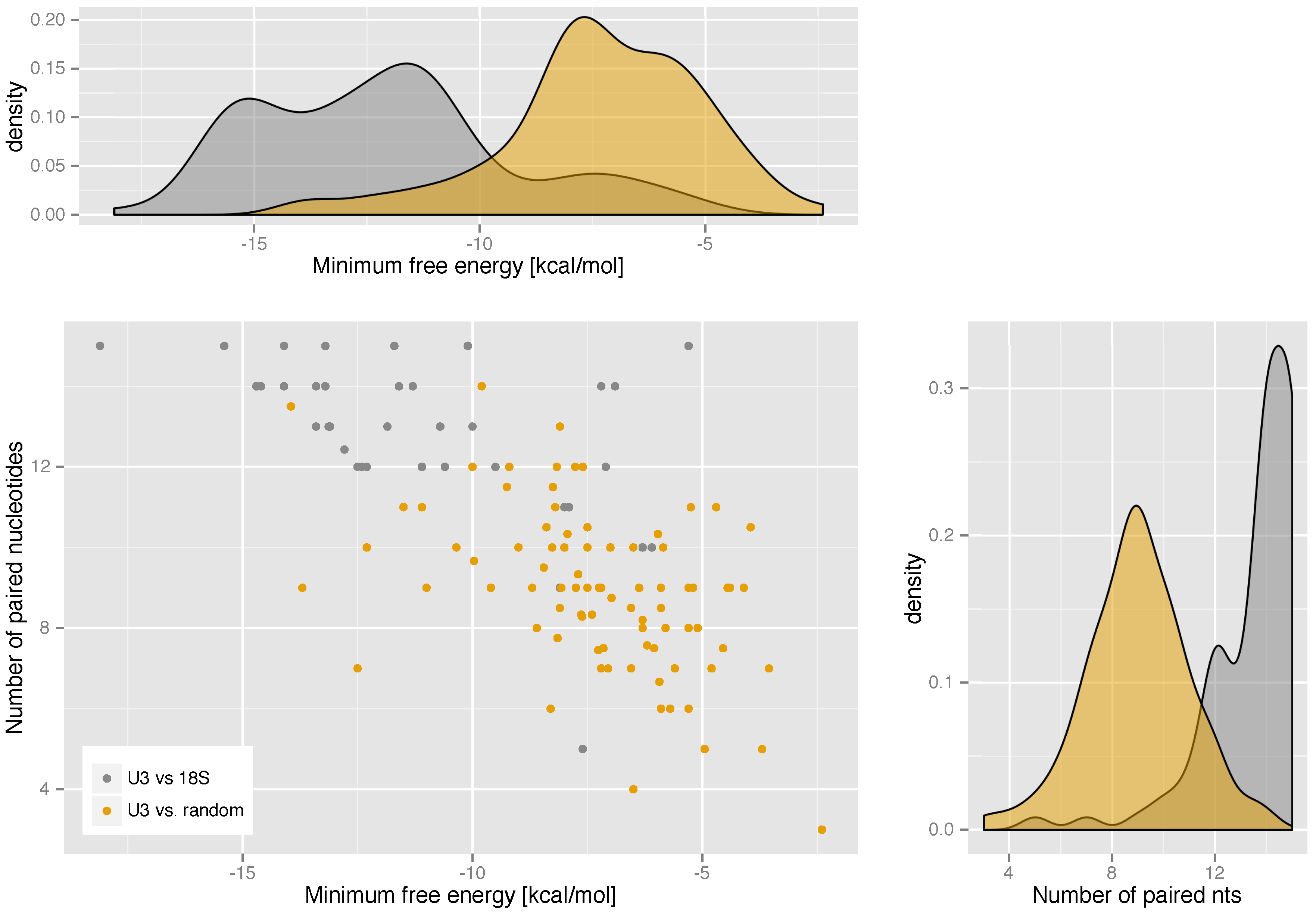
2.3. U3-Target Interactions
2.4. Characterization of Sequence Motifs
3. Results
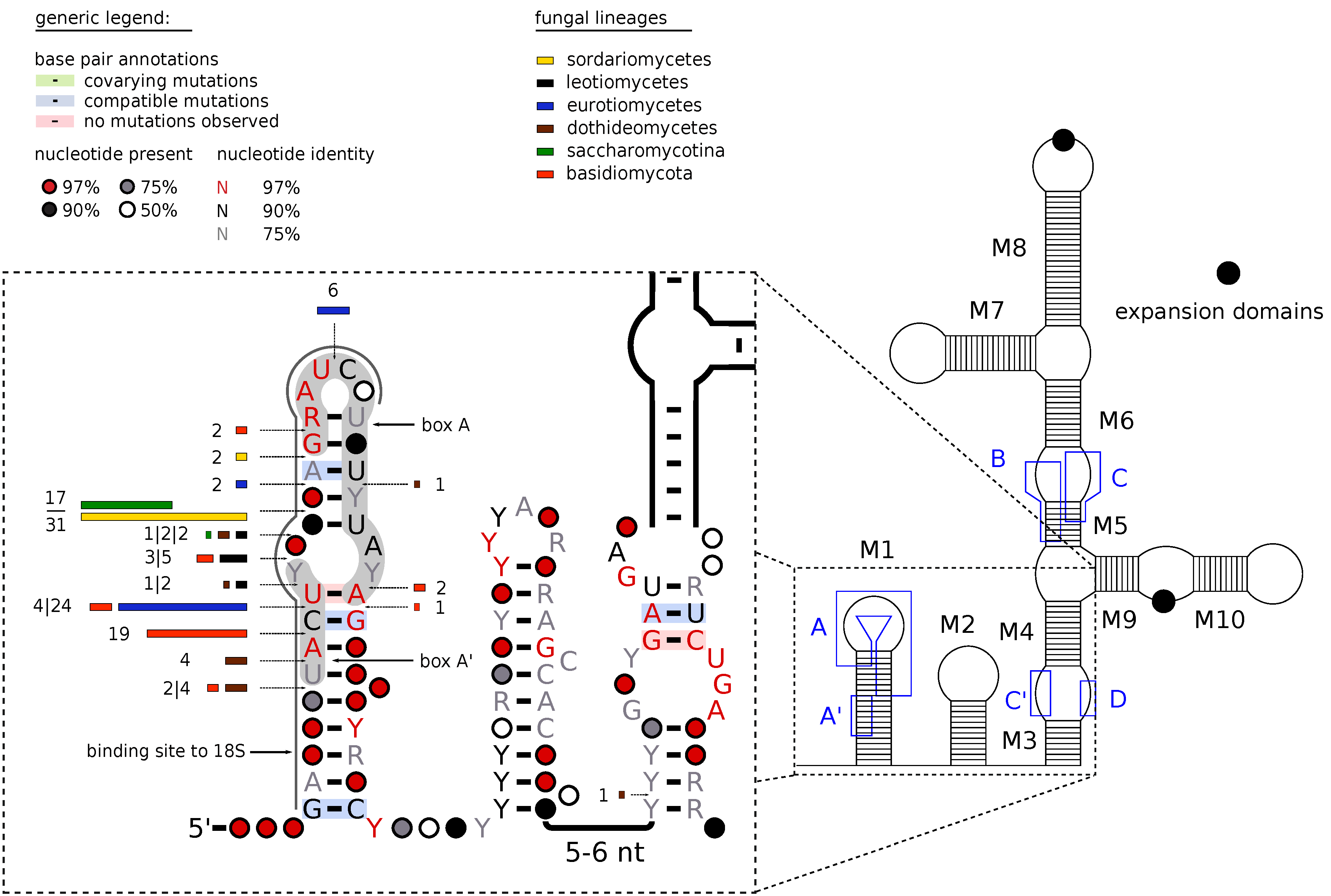
3.1. Sequence and Structure Motifs
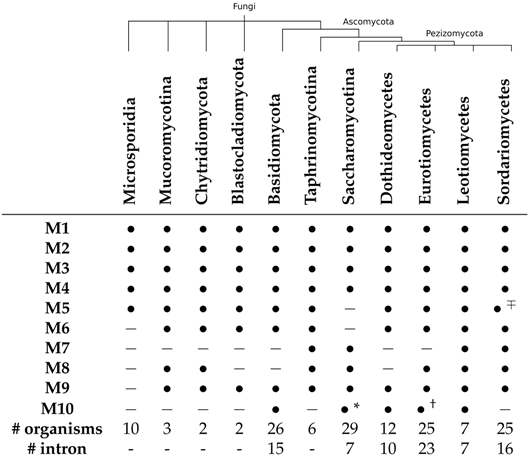
3.2. Introns within U3 snoRNA Genes
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
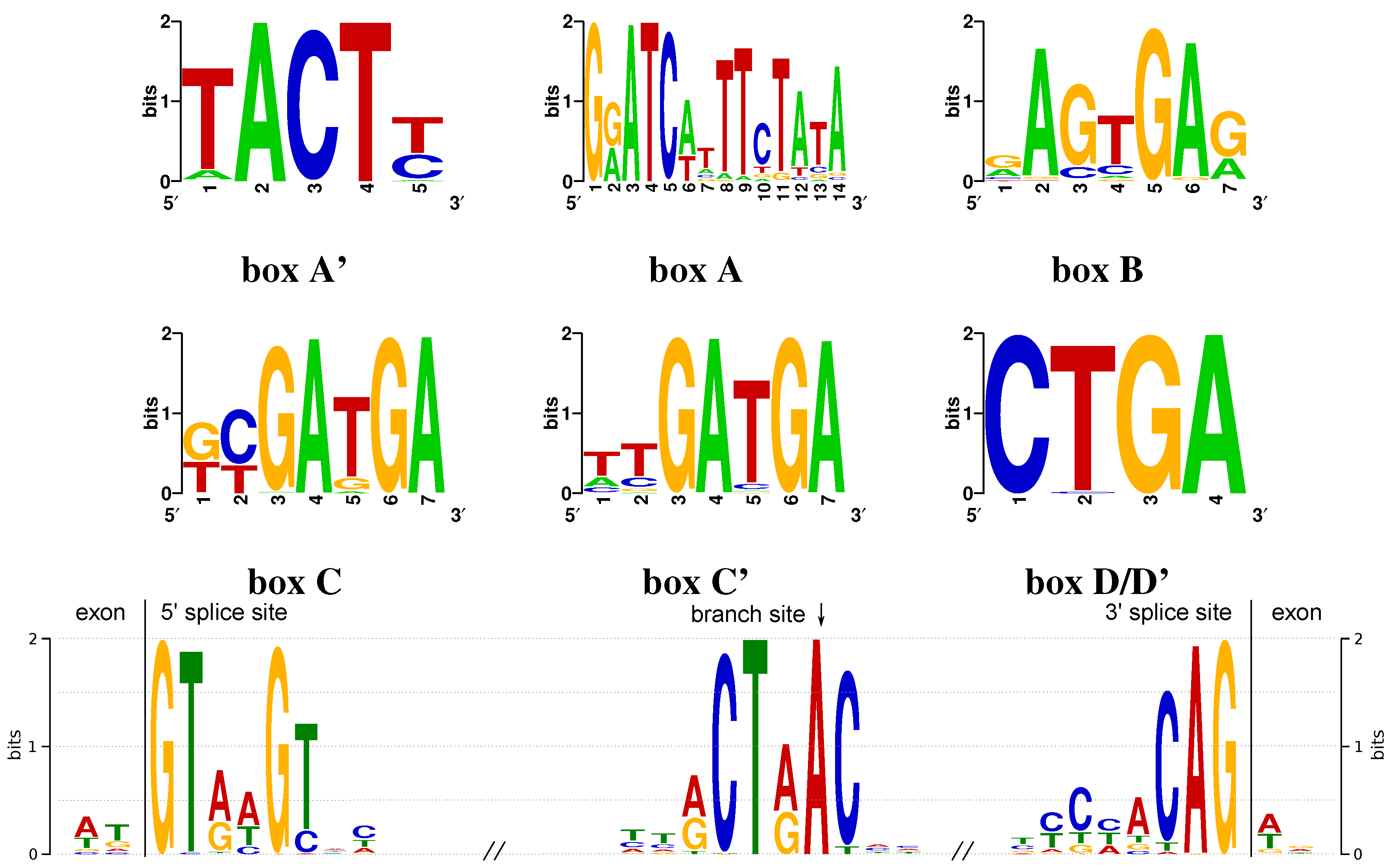
Appendix A. Conserved Box Motifs
References
- Henras, A.K.; Dez, C.; Henry, Y. RNA structure and function in C/D and H/ACA s(no)RNPs. Curr. Opin. Struct. Biol. 2004, 14, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Watkins, N.J.; Bohnsack, M.T. The box C/D and H/ACA snoRNPs: Key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA 2012, 3, 397–414. [Google Scholar] [CrossRef] [PubMed]
- Marz, M.; Stadler, P.F. Comparative Analysis of Eukaryotic U3 snoRNAs. RNA Biol. 2009, 6, 503–507. [Google Scholar] [CrossRef] [PubMed]
- Nabavi, S.; Nazar, R.N. U3 snoRNA promoter reflects the RNA’s function in ribosome biogenesis. Curr. Genet. 2008, 54, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Jia, D.; Cai, L.; He, H.; Skogerbø, G.; Li, T.; Aftab, M.N.; Chen, R. Systematic identification of non-coding RNA 2,2,7-trimethylguanosine cap structures in Caenorhabditis elegans. BMC Mol. Biol. 2007, 8, 86. [Google Scholar] [CrossRef] [PubMed]
- Samarsky, D.A.; Fournier, M.J. Functional mapping of the U3 small nucleolar RNA from the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1998, 18, 3431–3444. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Huang, Q.; Hui, Z.; Liang, Y.; Yu, C.; Qu, L. Identification, expression and functional analysis of U3 snoRNA genes from Neurospora crassa. Prog. Nat. Sci. 2009, 19, 167–172. [Google Scholar] [CrossRef]
- Dutca, L.M.; Gallagher, J.E.G.; Baserga, S.J. The initial U3 snoRNA:pre-rRNA base pairing interaction required for pre-18S rRNA folding revealed by in vivo chemical probing. Nucleic Acids Res. 2011, 39, 5164–5180. [Google Scholar] [CrossRef] [PubMed]
- Myslinski, E.; Ségault, V.; Branlant, C. An intron in the genes for U3 small nucleolar RNAs of the yeast Saccharomyces cerevisiae. Science 1990, 247, 1213–1216. [Google Scholar] [CrossRef] [PubMed]
- Selinger, D.A.; Porter, G.L.; Brennwald, P.J.; Wise, J.A. The two similarly expressed genes encoding U3 snRNA in Schizosaccharomyces pombe lack introns. Mol. Biol. Evol. 1992, 9, 297–308. [Google Scholar] [PubMed]
- Potashkin, J.; Frendewey, D. Splicing of the U6 RNA precursor is impaired in fission yeast pre-mRNA splicing mutants. Nucleic Acids Res 1989, 17, 7821–7831. [Google Scholar] [CrossRef] [PubMed]
- Tani, T.; Ohshima, Y. The gene for the U6 small nuclear RNA in fission yeast has an intron. Nature 1989, 337, 87–90. [Google Scholar] [CrossRef] [PubMed]
- Canzler, S.; Stadler, P.F.; Hertel, J. U6 snRNA intron insertion occurred multiple times during fungi evolution. RNA Biol. 2016, 13, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, Z.; Breaker, R.R. R2R—Software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinform. 2011, 12, 3. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nawrocki, E.P.; Kolbe, D.L.; Eddy, S.R. Infernal 1.0: Inference of RNA alignments. Bioinformatics 2009, 25, 1335–1337. [Google Scholar] [CrossRef] [PubMed]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef] [PubMed]
- Will, S.; Missal, K.; Hofacker, I.L.; Stadler, P.F.; Backofen, R. Inferring Non-Coding RNA Families and Classes by Means of Genome-Scale Structure-Based Clustering. PLoS Comp. Biol. 2007, 3, e65. [Google Scholar] [CrossRef] [PubMed]
- Washietl, S.; Hofacker, I.L.; Stadler, P.F. Fast and reliable prediction of noncoding RNAs. Proc. Natl. Acad. Sci. USA 2005, 102, 2454–2459. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, R.; Bernhart, S.H.; Höner zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar] [PubMed]
- Kupfer, D.M.; Drabenstot, S.D.; Buchanan, K.L.; Lai, H.; Zhu, H.; Dyer, D.W.; Roe, B.A.; Murphy, J.W. Introns and splicing elements of five diverse fungi. Eukaryot Cell 2004, 3, 1088–1100. [Google Scholar] [CrossRef] [PubMed]
- Liao, D. Concerted Evolution: Molecular Mechanisms and Biological Implications. Am. J. Hum. Genet. 1999, 64, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Fournier, R.; Brulé, F.; Ségault, V.; Mougin, A.; Branlant, C. U3 snoRNA genes with and without intron in the Kluyveromyces genus: Yeasts can accommodate great variations of the U3 snoRNA 3′-terminal domain. RNA 1998, 4, 285–302. [Google Scholar] [PubMed]
- Rep, M.; Duyvesteijn, R.G.; Gale, L.; Usgaard, T.; Cornelissen, B.J.; Ma, L.J.; Ward, T.J. The presence of GC-AG introns in Neurospora crassa and other euascomycetes determined from analyses of complete genomes: Implications for automated gene prediction. Genomics 2006, 87, 338–347. [Google Scholar] [CrossRef] [PubMed]
- Irimia, M.; Roy, S.W. Evolutionary convergence on highly-conserved 3′ intron structures in intron-poor eukaryotes and insights into the ancestral eukaryotic genome. PLoS Genet. 2008, 4, e1000148. [Google Scholar] [CrossRef] [PubMed]
- Mertins, P.; Gallwitz, D. Nuclear pre-mRNA splicing in the fission yeast Schizosaccharomyces pombe strictly requires an intron-contained, conserved sequence element. EMBO J. 1987, 6, 1757–1763. [Google Scholar] [PubMed]
- Rogozin, I.B.; Lyons-Weiler, J.; Koonin, E.V. Intron sliding in conserved gene families. Trends Genet. 2000, 16, 430–432. [Google Scholar] [CrossRef]
- Puchta, O.; Cseke, B.; Czaja, H.; Tollervey, D.; Sanguinetti, G.; Kudla, G. Network of epistatic interactions within a yeast snoRNA. Science 2016, 352, 840–844. [Google Scholar] [CrossRef] [PubMed]
- Hooks, K.B.; Delneri, D.; Griffiths-Jones, S. Intron evolution in Saccharomycetaceae. Genome Biol. Evol. 2014, 6, 2543–2556. [Google Scholar] [CrossRef] [PubMed]
 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Canzler, S.; Stadler, P.F.; Hertel, J. Evolution of Fungal U3 snoRNAs: Structural Variation and Introns. Non-Coding RNA 2017, 3, 3. https://doi.org/10.3390/ncrna3010003
Canzler S, Stadler PF, Hertel J. Evolution of Fungal U3 snoRNAs: Structural Variation and Introns. Non-Coding RNA. 2017; 3(1):3. https://doi.org/10.3390/ncrna3010003
Chicago/Turabian StyleCanzler, Sebastian, Peter F. Stadler, and Jana Hertel. 2017. "Evolution of Fungal U3 snoRNAs: Structural Variation and Introns" Non-Coding RNA 3, no. 1: 3. https://doi.org/10.3390/ncrna3010003






