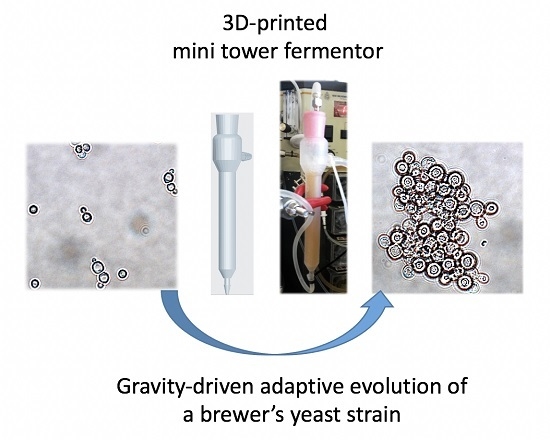
Gravity-Driven Adaptive Evolution of an Industrial Brewer’s Yeast Strain towards a Snowflake Phenotype in a 3D-Printed Mini Tower Fermentor
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Media
2.2. Determination of Cell Concentration and Growth Rate
2.3. Flocculation Assay
2.4. Biocompatibility Assay
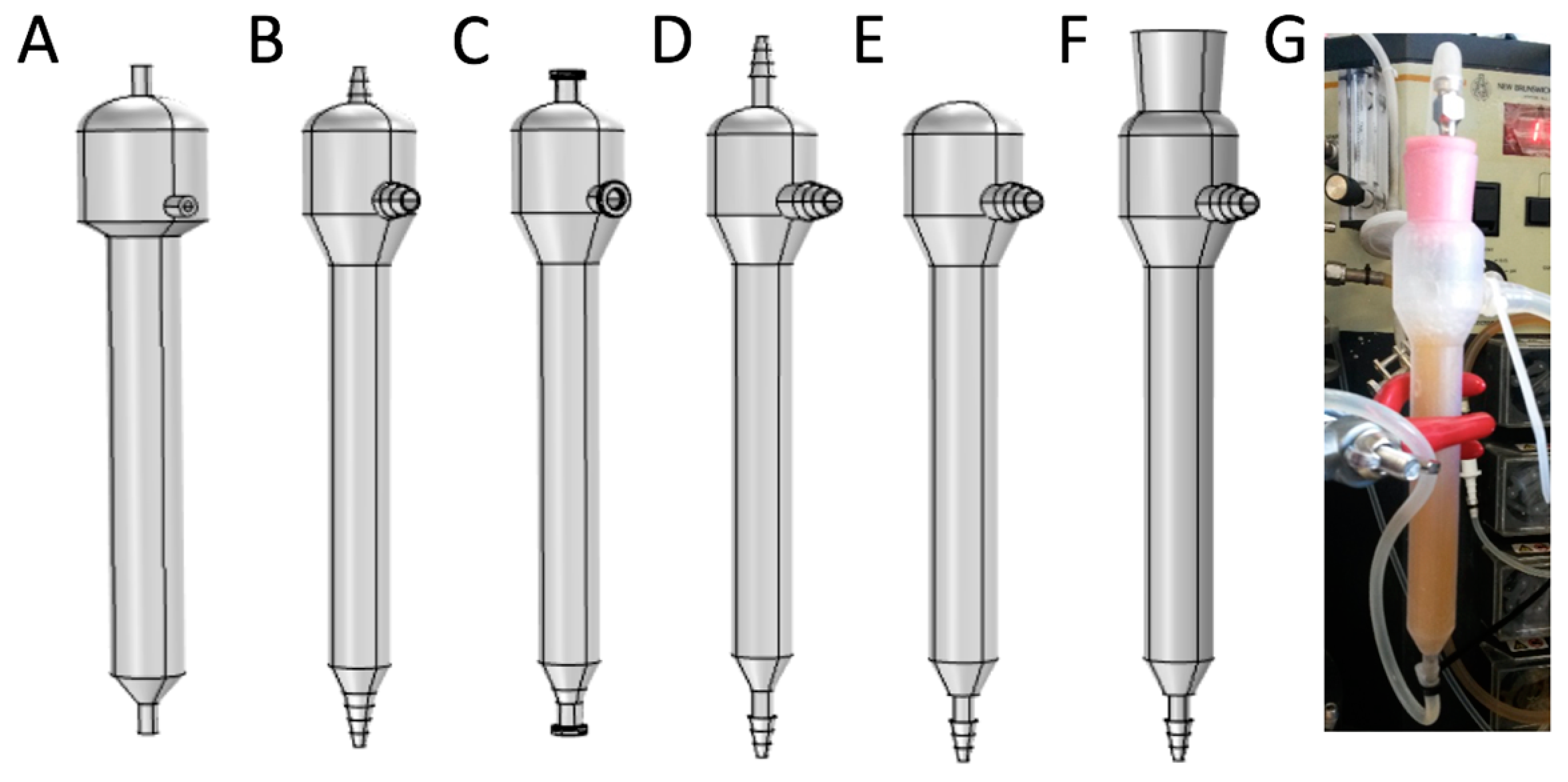
2.5. Mini Tower Fermentor Design and Construction
2.6. Mini Tower Fermentor Mathematical Modelling
2.7. Experimental Set-Up
3. Results
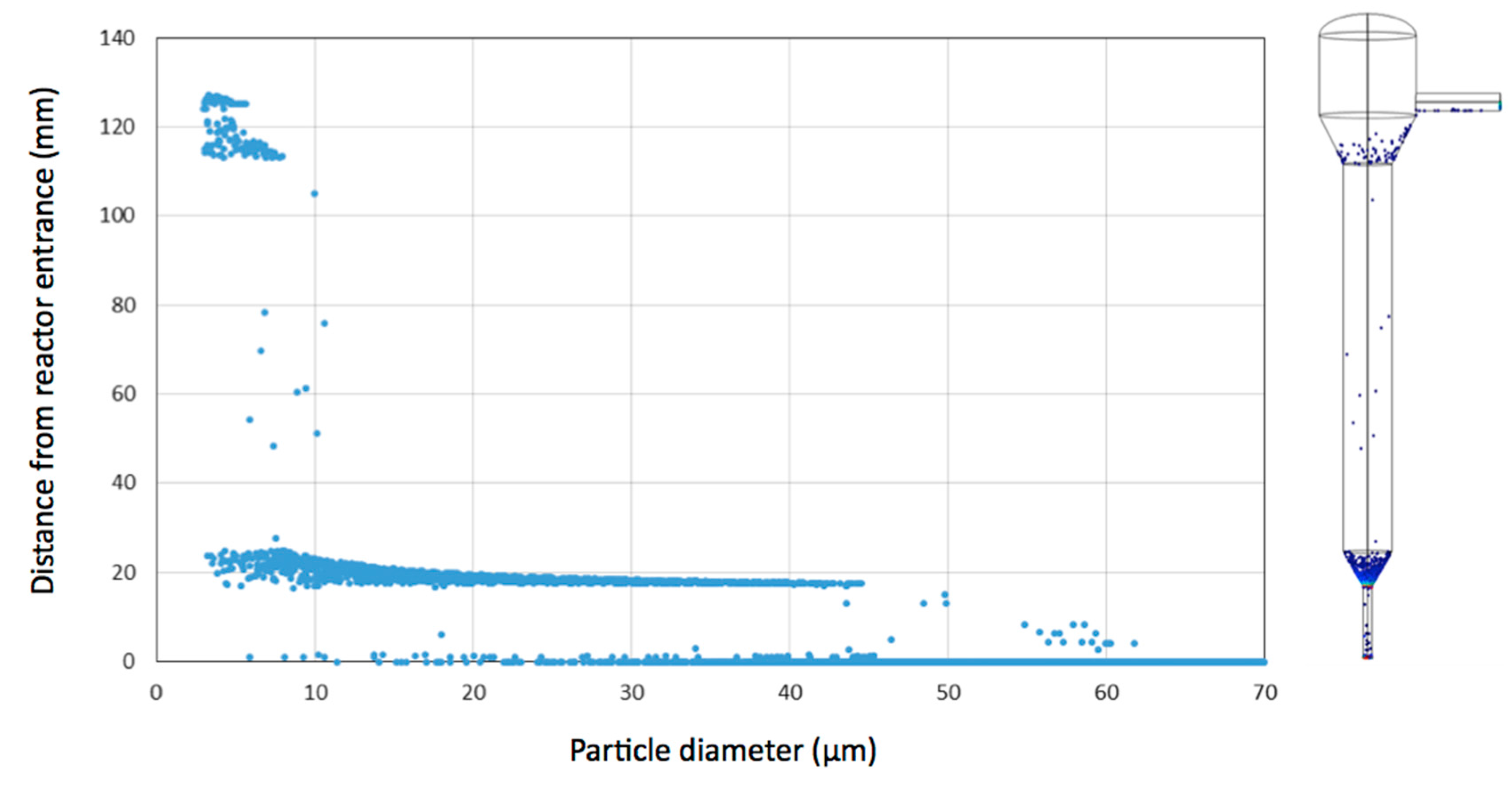
3.1. Simulation of Single Cell and Cell Aggregate Behaviour in the Mini Tower Fermentor
3.2. Biocompatibility Assay
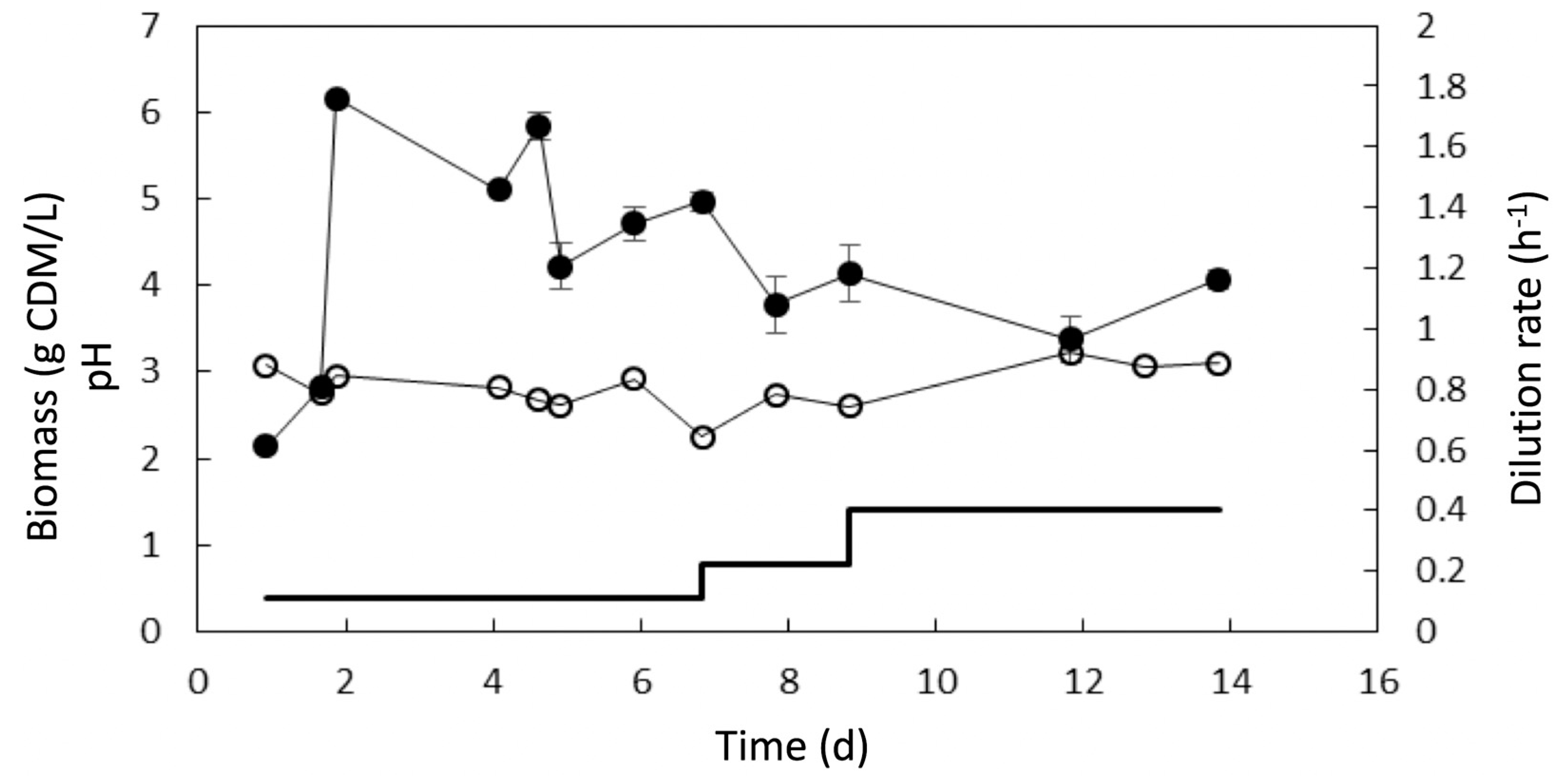
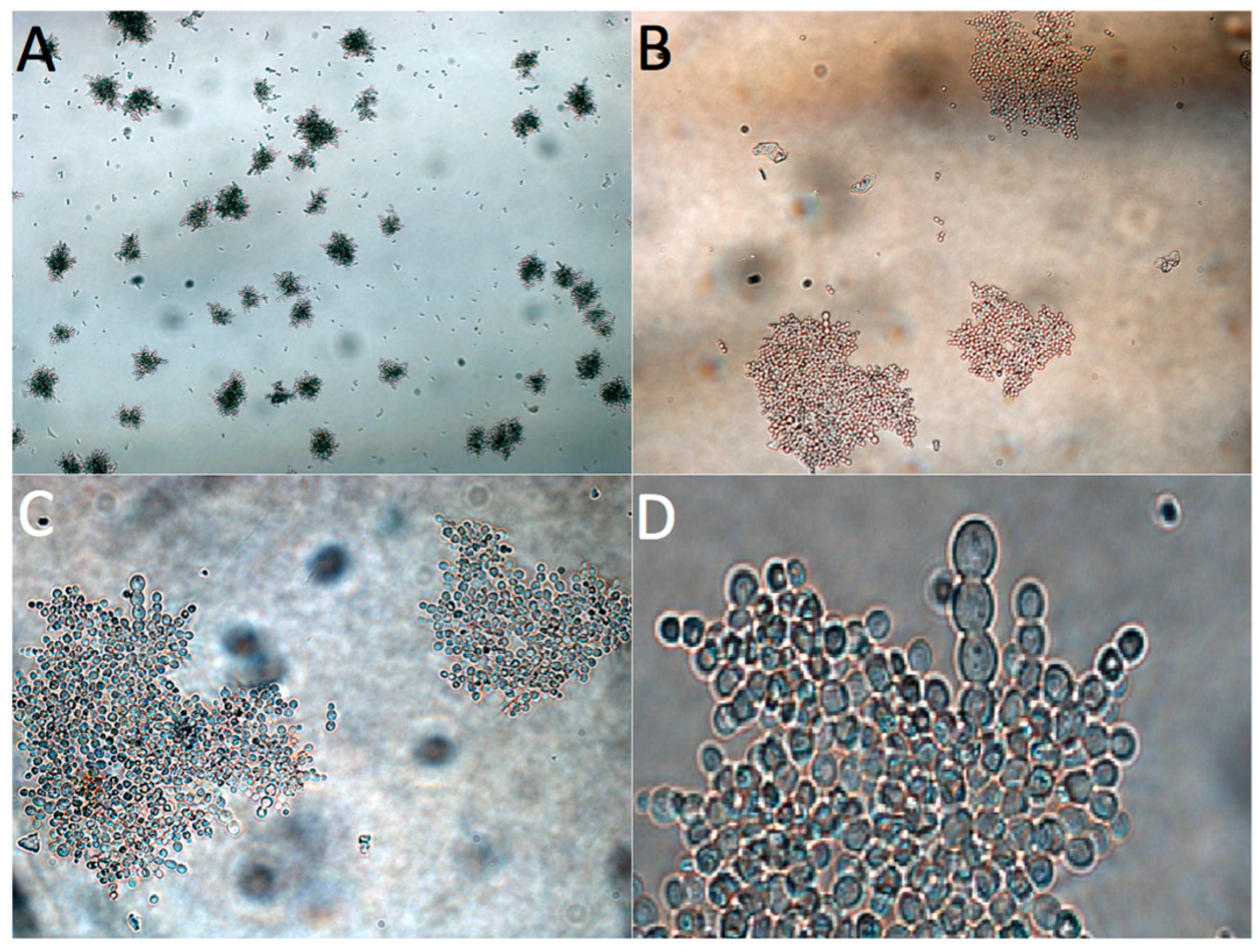
3.3. Adaptive Evolution during Continuous Operation
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A
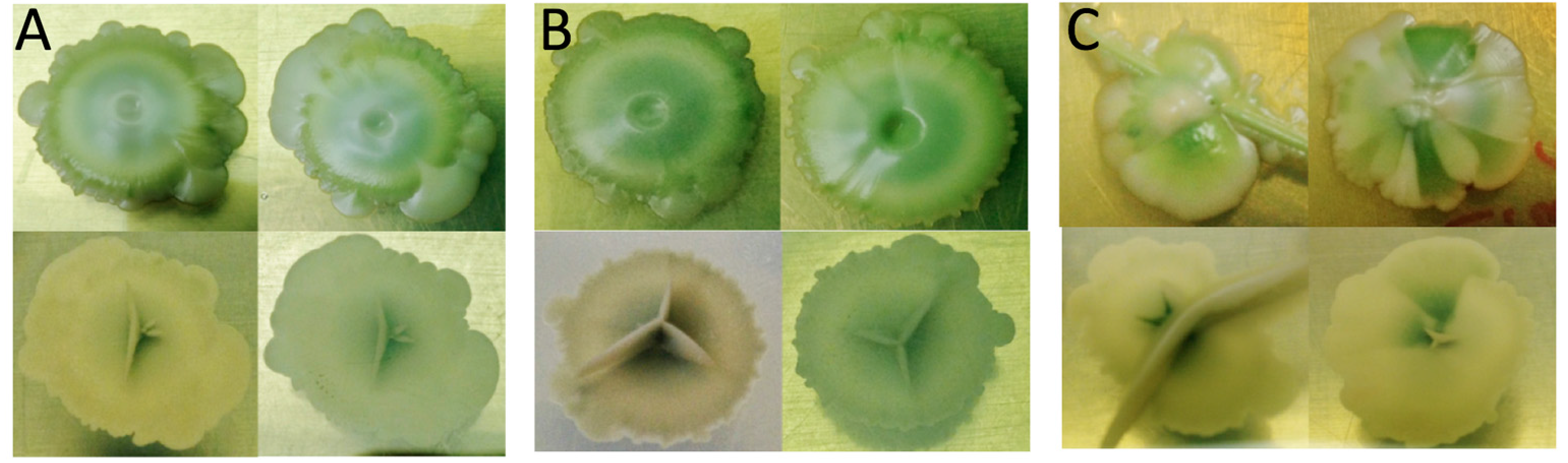
 ) of the methacrylic acid ester resin printed material during 1 day of growth.
) of the methacrylic acid ester resin printed material during 1 day of growth.
 ) of the methacrylic acid ester resin printed material during 1 day of growth.
) of the methacrylic acid ester resin printed material during 1 day of growth.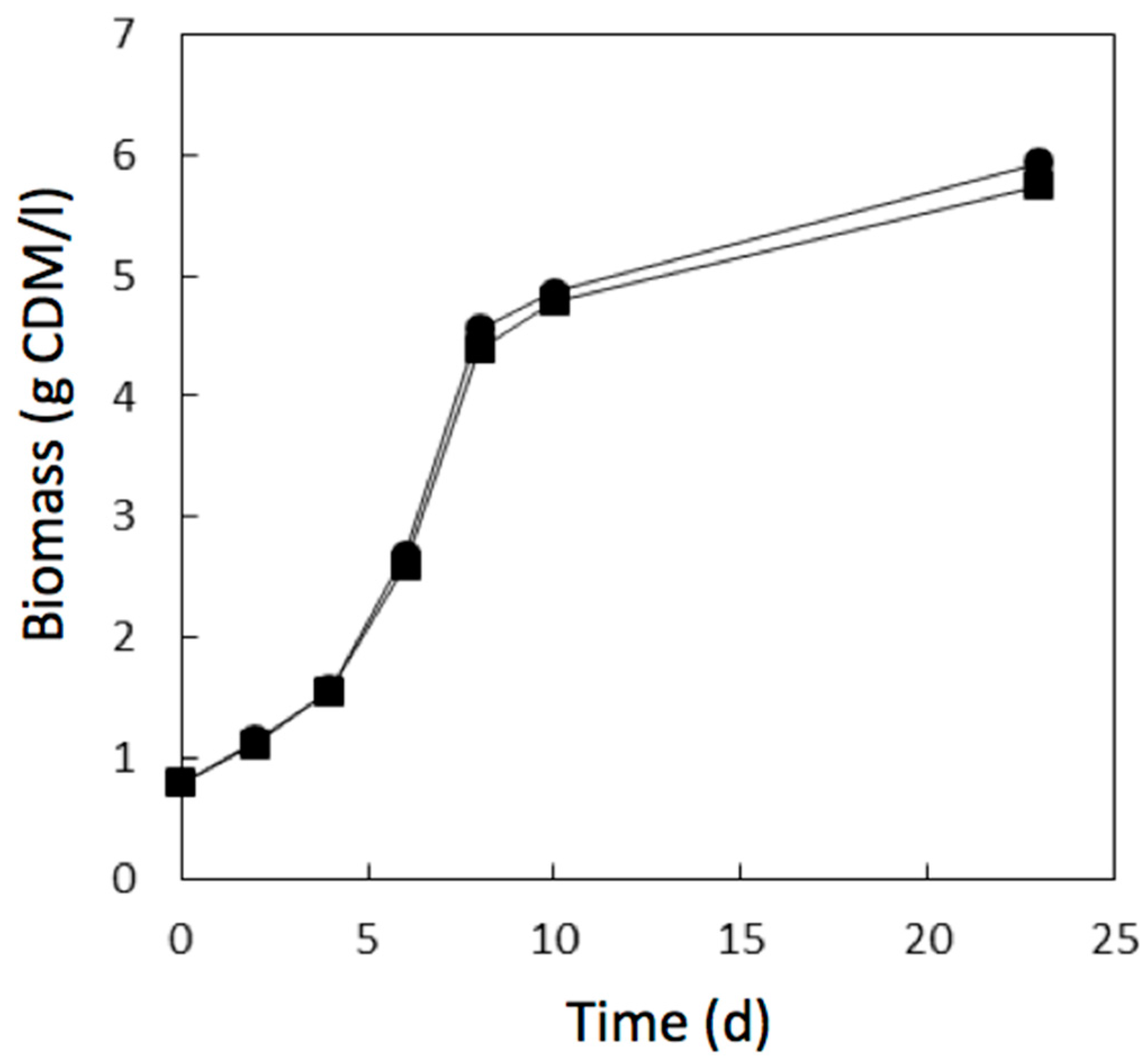
References
- Dequin, S. The potential of genetic engineering for improving brewing, wine-making and baking yeasts. Appl. Microbiol. Biotechnol. 2001, 56, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Saerens, S.M.; Duong, C.T.; Nevoigt, E. Genetic improvement of brewer’s yeast: Current state, perspectives and limits. Appl. Microbiol. Biotechnol. 2010, 86, 1195–1212. [Google Scholar] [CrossRef] [PubMed]
- Conrad, T.M.; Lewis, N.E.; Palsson, B.Ø. Microbial laboratory evolution in the era of genome-scale science. Mol. Syst. Biol. 2011, 7, 509. [Google Scholar] [CrossRef] [PubMed]
- Portnoy, V.A.; Bezdan, D.; Zengler, K. Adaptive laboratory evolution—Harnessing the power of biology for metabolic engineering. Curr. Opin. Biotechnol. 2011, 22, 590–594. [Google Scholar] [CrossRef] [PubMed]
- Dragosits, M.; Mattanovich, D. Adaptive laboratory evolution—Principles and applications for biotechnology. Microb. Cell Fact. 2013, 12, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibson, B.; Liti, G. Saccharomyces pastorianus: Genomic insights inspiring innovation for industry. Yeast 2015, 32, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Blieck, L.; Toye, G.; Dumortier, F.; Verstrepen, K.J.; Delvaux, F.R.; Thevelein, J.M.; van Dijck, P. Isolation and characterization of brewer’s yeast variants with improved fermentation performance under high-gravity conditions. Appl. Environ. Microbiol. 2007, 73, 815–824. [Google Scholar] [CrossRef] [PubMed]
- Huuskonen, A.; Markkula, T.; Vidgren, V.; Lima, L.; Mulder, L.; Geurts, W.; Walsh, M.; Londesborough, J. Selection from industrial lager yeast strains of variants with improved fermentation performance in very-high-gravity worts. Appl. Environ. Microbiol. 2010, 76, 1563–1573. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zhao, H.; Li, H.; Zhang, Q.; Lei, H.; Zhao, M. Selection of Saccharomyces pastorianus variants with improved fermentation performance under very high gravity wort conditions. Biotechnol. Lett. 2012, 34, 365–370. [Google Scholar] [CrossRef] [PubMed]
- James, T.C.; Usher, J.; Campbell, S.; Bond, U. Lager yeasts possess dynamic genomes that undergo rearrangements and gene amplification in response to stress. Curr. Genet. 2008, 53, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Ekberg, J.; Rautio, J.; Mattinen, L.; Vidgren, V.; Londesborough, J.; Gibson, B.R. Adaptive evolution of the lager brewing yeast Saccharomyces pastorianus for improved growth under hyperosmotic conditions and its influence on fermentation performance. FEMS Yeast Res. 2013, 13, 335–349. [Google Scholar] [CrossRef] [PubMed]
- Strejc, J.; Siříštova, L.; Karabín, M.; Almeida e Silva, J.B.; Brányik, T. Production of alcohol-free beer with elevated amounts of flavouring compounds using lager yeast mutants. J. Inst. Brew. 2013, 119, 149–155. [Google Scholar] [CrossRef]
- Cadière, A.; Ortiz-Julien, A.; Camarasa, C.; Dequin, S. Evolutionary engineered Saccharomyces cerevisiae wine yeast strains with increased in vivo flux through the pentose phosphate pathway. Metab. Eng. 2011, 13, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Cadière, A.; Aguera, E.; Caillé, S.; Ortiz-Julien, A.; Dequin, S. Pilot-scale evaluation the enological traits of a novel, aromatic wine yeast strain obtained by adaptive evolution. Food Microbiol. 2012, 32, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Dunham, M.J.; Kerr, E.O.; Miller, A.W.; Payen, C. Chemostat culture for yeast physiology and experimental evolution. Cold Spring Harb. Protoc. 2016. [Google Scholar] [CrossRef]
- Miller, A.W.; Befort, C.; Kerr, E.O.; Dunham, M.J. Design and use of multiplexed chemostat arrays. J. Vis. Exp. 2013, 72, e50262. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Wittmann, C.; Heinzle, E. Minibioreactors. Biotechnol. Lett. 2004, 26, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Betts, J.I.; Baganz, F. Miniature bioreactors: Current practices and future opportunities. Microb. Cell Fact. 2006, 5, 21. [Google Scholar] [CrossRef] [PubMed]
- Hortsch, R.; Weuster-Botz, D. Milliliter-scale stirred tank reactors for the cultivation of microorganisms. Adv. Appl. Microbiol. 2010, 73, 61–82. [Google Scholar] [PubMed]
- Tuomi, J.; Paloheimo, K.S.; Vehviläinen, J.; Björkstrand, R.; Salmi, M.; Huotilainen, E.; Kontio, R.; Rouse, S.; Gibson, I.; Mäkitie, A.A. A novel classification and online platform for planning and documentation of medical applications of additive manufacturing. Surg. Innov. 2014, 21, 553–559. [Google Scholar] [CrossRef] [PubMed]
- Mironov, V.; Boland, T.; Trusk, T.; Forgacs, G.; Markwald, R.R. Organ printing: Computer-aided jet-based 3D tissue engineering. Trends Biotechnol. 2003, 21, 157–161, Erratum in: Trends Biotechnol. 2004, 22, 265. [Google Scholar] [CrossRef]
- Mironov, V.; Kasyanov, V.; Markwald, R.R. Organ printing: From bioprinter to organ biofabrication line. Curr. Opin. Biotechnol. 2011, 22, 667–673. [Google Scholar] [CrossRef] [PubMed]
- Leukers, B.; Gülkan, H.; Irsen, S.H.; Milz, S.; Tille, C.; Schieker, M.; Seitz, H. Hydroxyapatite scaffolds for bone tissue engineering made by 3D printing. J. Mater. Sci. Mater. Med. 2005, 16, 1121–1124. [Google Scholar] [CrossRef] [PubMed]
- Salmi, M. Possibilities of preoperative medical models made by 3D printing or additive manufacturing. J. Med. Eng. 2016, 2016, 6191526. [Google Scholar] [CrossRef] [PubMed]
- Sears, N.A.; Seshadri, D.R.; Dhavalikar, P.S.; Cosgriff-Hernandez, E. A review of three-dimensional printing in tissue engineering. Tissue Eng. Part B Rev. 2016, 22, 298–310. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Fan, S.Q.; Shen, Y.; Yang, J.X.; Yan, P.; Chen, Y.P.; Li, J.; Guo, J.S.; Duan, X.M.; Fang, F.; et al. A novel bio-carrier fabricated using 3D printing technique for wastewater treatment. Sci. Rep. 2015, 5, 12400. [Google Scholar] [CrossRef] [PubMed]
- Gross, B.C.; Erkal, J.L.; Lockwood, S.Y.; Chen, C.; Spence, D.M. Evaluation of 3D printing and its potential impact on biotechnology and the chemical sciences. Anal. Chem. 2014, 86, 3240–3253. [Google Scholar] [CrossRef] [PubMed]
- Melchels, F.P.; Feijen, J.; Grijpma, D.W. A review on stereolithography and its applications in biomedical engineering. Biomaterials 2010, 31, 6121–6130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boulton, C.; Quain, D. Fermentation systems. In Brewing Yeast and Fermentation; Blackwell Science Ltd.: Oxford, UK, 2001; pp. 260–372. [Google Scholar]
- Klopper, W.J.; Roberts, R.H.; Royston, M.G.; Ault, R.G. Continuous fermentation in a tower fermenter. Proc. Eur. Brew. Conv. 1965, 238–259. [Google Scholar]
- Ault, R.G.; Hampton, A.N.; Newton, R.; Roberts, R.H. Biological and biochemical aspects of tower fermentation. J. Inst. Brew. 1969, 75, 260–277. [Google Scholar] [CrossRef]
- Van Mulders, S.E.; Ghequire, M.; Daenen, L.; Verbelen, P.J.; Verstrepen, K.J.; Delvaux, F.R. Flocculation gene variability in industrial brewer’s yeast strains. Appl. Microbiol. Biotechnol. 2010, 88, 1321–1331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brachmann, C.B.; Davies, A. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 1998, 14, 115–132. [Google Scholar] [CrossRef]
- Van Mulders, S.E.; Christianen, E.; Saerens, S.M.G.; Daenen, L.; Verbelen, P.J.; Willaert, R.; Verstrepen, K.J.; Delvaux, F.R. Phenotypic diversity of Flo protein family-mediated adhesion in Saccharomyces cerevisiae. FEMS Yeast Res. 2009, 9, 178–190. [Google Scholar] [CrossRef] [PubMed]
- Richards, M. The use of giant-colony morphology for the differentiation of brewing yeasts. J. Inst. Brew. 1967, 73, 162–166. [Google Scholar] [CrossRef]
- Pallmann, C.L.; Brown, J.A.; Olineka, T.L.; Cocolin, L.; Mills, D.A.; Bisson, L.F. Use of WL medium to profile native flora fermentations. Am. J. Enol. Vitic. 2001, 52, 198–203. [Google Scholar]
- D’Hautcourt, O.; Smart, K.A. Measurement of brewing yeast flocculation. J. Am. Soc. Brew. Chem. 1999, 57, 123–128. [Google Scholar]
- Den Blanken, J.G. The tower fermenter for lager beer production. Brew. Gardian 1974, 35–39. [Google Scholar]
- Seddon, A.W. Continuous tower fermentation—Experiences in establishing large scale commercial production. Brewer 1976, 72–78. [Google Scholar]
- Haddad, S.A.; Lindegren, C.C. A method for determining the weight of an individual yeast cell. Appl. Microbiol. 1953, 1, 153–156. [Google Scholar] [PubMed]
- Davis, R.H.; Hunt, T.P. Modeling and measurement of yeast flocculation. Biotechnol. Prog. 1986, 2, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Fontana, A.; Bore, C.; Ghommidh, C.; Guiraud, J.P. Structure and sucrose hydrolysis activity of Saccharomyces cerevisiae aggregates. Biotechnol. Bioeng. 1992, 40, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Van Hamersveld, E.H.; Van Der Lans, R.G.; Luyben, K.C. Quantification of brewers’ yeast flocculation in a stirred tank: Effect of physical parameters on flocculation. Biotechnol. Bioeng. 1997, 56, 190–200. [Google Scholar] [CrossRef]
- Ratcliff, W.C.; Denison, R.F.; Borrello, M.; Travisano, M. Experimental evolution of multicellularity. Proc. Natl. Acad. Sci. USA 2012, 109, 1595–1600. [Google Scholar] [CrossRef] [PubMed]
- Oud, B.; Guadalupe-Medina, V.; Nijkamp, J.F.; de Ridder, D.; Pronk, J.T.; van Maris, A.J.; Daran, J.M. Genome duplication and mutations in ACE2 cause multicellular, fast-sedimenting phenotypes in evolved Saccharomyces cerevisiae. Proc. Natl. Acad. Sci USA 2013, 110, E4223–E4231. [Google Scholar] [CrossRef] [PubMed]
- Ratcliff, W.C.; Fankhauser, J.D.; Rogers, D.W.; Greig, D.; Travisano, M. Origins of multicellular evolvability in snowflake yeast. Nat. Commun. 2015, 6, 6102. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Styles, C.A.; Feng, Q.; Fink, G.R. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. USA 2000, 97, 12158–12163. [Google Scholar] [CrossRef] [PubMed]
- Goossens, K.V.; Willaert, R.G. The N-terminal domain of the Flo11 protein from Saccharomyces cerevisiae is an adhesin without mannose-binding activity. FEMS Yeast Res. 2012, 12, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Kraushaar, T.; Brückner, S.; Veelders, M.; Rhinow, D.; Schreiner, F.; Birke, R.; Pagenstecher, A.; Mösch, H.U.; Essen, L.O. Interactions by the fungal Flo11 adhesin depend on a fibronectin Type III-like adhesin domain girdled by aromatic bands. Structure 2015, 23, 1005–1017. [Google Scholar] [CrossRef] [PubMed]
- Mill, P.J. The nature of the interactions between flocculent cells in the flocculation of Saccharomyces cerevisiae. J. Gen. Microbiol. 1964, 35, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Stratford, M.; Keenan, M.H. Yeast flocculation: Quantification. Yeast 1988, 4, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Weiss, E.L. Mitotic exit and separation of mother and daughter cells. Genetics 2012, 192, 1165–1202. [Google Scholar] [CrossRef] [PubMed]
- King, L.; Butler, G. Ace2p, a regulator of CTS1 (chitinase) expression, affects pseudohyphal production in Saccharomyces cerevisiae. Curr. Genet. 1998, 34, 183–191. [Google Scholar] [CrossRef] [PubMed]
- O’Conallain, C.; Doolin, M.T.; Taggart, C.; Thornton, F.; Butler, G. Regulated nuclear localisation of the yeast transcription factor Ace2p controls expression of chitinase (CTS1) in Saccharomyces cerevisiae. Mol. Gen. Genet. 1999, 262, 275–282. [Google Scholar] [CrossRef] [PubMed]
- McBride, H.J.; Yu, Y.; Stillman, D.J. Distinct regions of the Swi5 and Ace2 transcription factors are required for specific gene activation. J. Biol. Chem. 1999, 274, 21029–21036. [Google Scholar] [CrossRef] [PubMed]
- Bidlingmaier, S.; Weiss, E.L.; Seidel, C.; Drubin, D.G.; Snyder, M. The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001, 21, 2449–2462. [Google Scholar] [CrossRef] [PubMed]
- Voth, W.P.; Olsen, A.E.; Sbia, M.; Freedman, K.H.; Stillman, D.J. ACE2, CBK1, and BUD4 in budding and cell separation. Eukaryot Cell 2005, 4, 1018–1028. [Google Scholar] [CrossRef] [PubMed]
- Versele, M.; Thevelein, J.M. Lre1 affects chitinase expression, trehalose accumulation and heat resistance through inhibition of the Cbk1 protein kinase in Saccharomyces cerevisiae. Mol. Microbiol. 2001, 41, 1311–1326. [Google Scholar] [CrossRef] [PubMed]
- Weiss, E.L.; Kurischko, C.; Zhang, C.; Shokat, K.; Drubin, D.G.; Luca, F.C. The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of Ace2p transcription factor. J. Cell Biol. 2002, 158, 885–900. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Conjaerts, A.; Willaert, R.G. Gravity-Driven Adaptive Evolution of an Industrial Brewer’s Yeast Strain towards a Snowflake Phenotype in a 3D-Printed Mini Tower Fermentor. Fermentation 2017, 3, 4. https://doi.org/10.3390/fermentation3010004
Conjaerts A, Willaert RG. Gravity-Driven Adaptive Evolution of an Industrial Brewer’s Yeast Strain towards a Snowflake Phenotype in a 3D-Printed Mini Tower Fermentor. Fermentation. 2017; 3(1):4. https://doi.org/10.3390/fermentation3010004
Chicago/Turabian StyleConjaerts, Andreas, and Ronnie G. Willaert. 2017. "Gravity-Driven Adaptive Evolution of an Industrial Brewer’s Yeast Strain towards a Snowflake Phenotype in a 3D-Printed Mini Tower Fermentor" Fermentation 3, no. 1: 4. https://doi.org/10.3390/fermentation3010004