Why Are Weissella spp. Not Used as Commercial Starter Cultures for Food Fermentation?
Abstract
:1. Introduction
2. Weissella Genus
2.1. Phenotypic and Genetic Characteristics of Weissella spp.
2.2. Taxonomical Position and Specific Traits
3. Occurrence of Weissella in Spontaneous Fermentation
3.1. General Occurrence
3.2. Fruits and Vegetables-Based Products
3.3. Dairy Fermented Foods
3.4. Meat and Fish-Based Products
3.5. Starchy and Cereal-Based Products
4. Spontaneous Versus Started Fermentation
4.1. Role of Starters in Food Fermentation
4.2. Expected Characteristics of Starters
4.3. Non-Expected Characteristics of Starters
4.4. Improvement of Starters
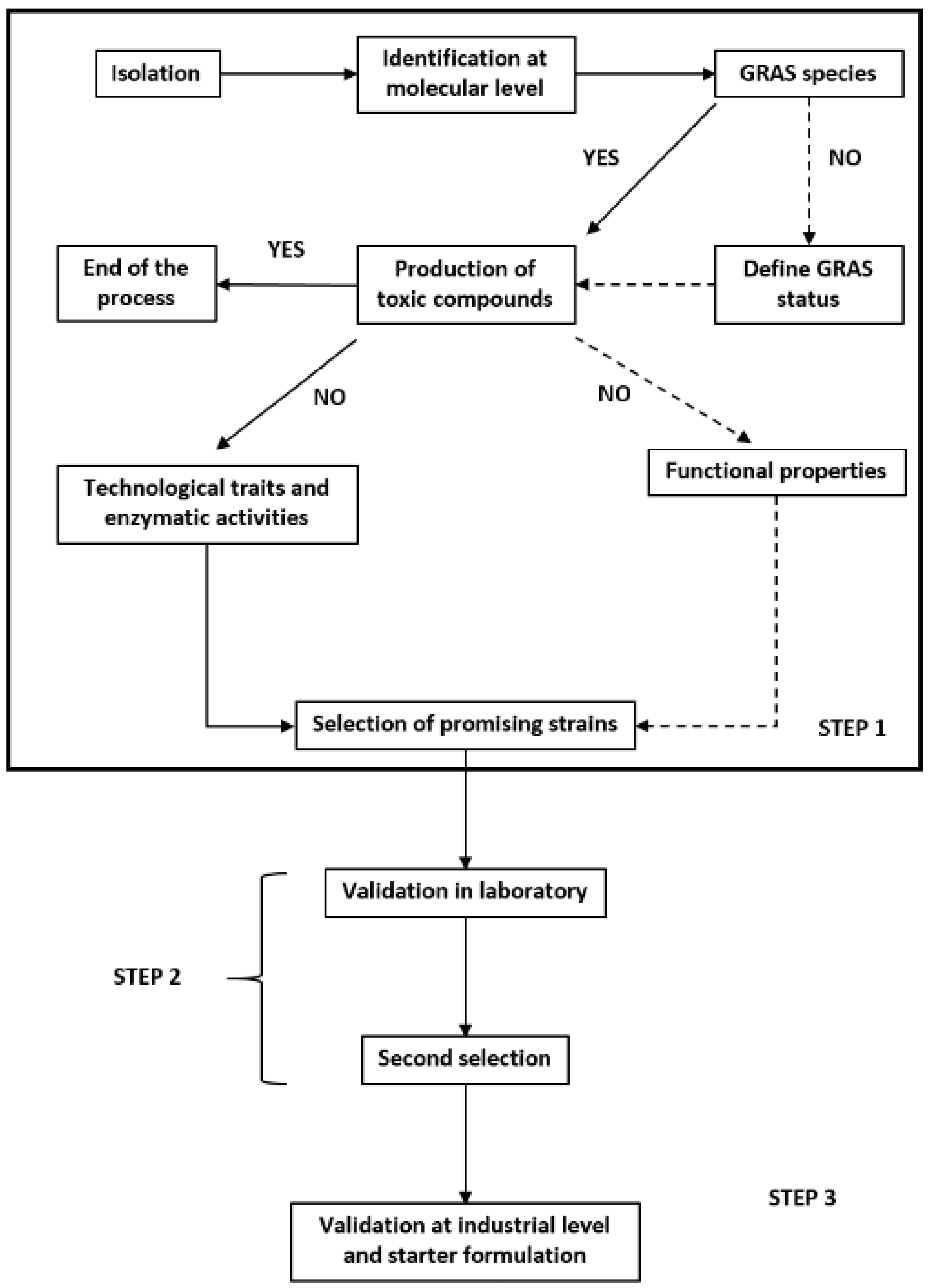
5. Investigation into Using Weissella spp. as a Starter
5.1. Acidification and Growth Performance
5.2. Production of Exopolysaccharides
5.3. Production of Antimicrobial Substances
5.4. Probiotic Aspects
5.5. Role in Bioavailability and Antioxidant Activities
6. Possible Constraints for the Use of Weissella spp. as Starters
6.1. Biogenic Amine Production of Weissella
6.2. Antibiotic Resistance Profile of Weissella
6.3. Infections Associated with Weissella
6.4. Bacteriophage Infection
6.5. Regulations
7. Concluding Remarks
Conflicts of Interest
References
- Ross, P.R.; Morgan, S.; Hill, C. Preservation and fermentation: Past, present and future. Int. J. Food Microbiol. 2002, 79, 3–16. [Google Scholar] [CrossRef]
- Tamang, J.P.; Watanabe, K.; Holzapfel, W.H. Review: Diversity of microorganisms in global fermented foods and beverages. Front. Microbiol. 2016, 7, 377. [Google Scholar] [CrossRef] [PubMed]
- Buckenhüskes, H.J. Selection criteria for lactic acid bacteria to be used as starter cultures for various food commodities. FEMS Microbiol. Rev. 1993, 12, 253–271. [Google Scholar] [CrossRef]
- Caplice, E.; Fitzgerald, G.F. Food fermentations: Role of microorganisms in food production and preservation. Int. J. Food Microbiol. 1999, 50, 131–149. [Google Scholar] [CrossRef]
- Stiles, M.E.; Holzapfel, W.H. Lactic acid bacteria of foods and their current taxonomy. Int. J. Food Microbiol. 1997, 36, 1–29. [Google Scholar] [CrossRef]
- Brandt, M.J. Starter cultures for cereal based foods. Food Microbiol. 2014, 37, 41–43. [Google Scholar] [CrossRef] [PubMed]
- Di Cagno, R.; Coda, R.; De Angelis, M.; Gobbetti, M. Exploitation of vegetables and fruits through lactic acid fermentation. Food Microbiol. 2013, 33, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Leroy, F.; De Vuyst, L. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci. Technol. 2004, 15, 67–78. [Google Scholar] [CrossRef]
- Amari, M.; Arango, L.F.G.; Gabriel, V.; Robert, H.; Morel, S.; Moulis, C.; Gabriel, B.; Remaud-Siméon, M.; Fontagné-Faucher, C. Characterization of a novel dextransucrase from Weissella confusa isolated from sourdough. Appl. Microbiol. Biotechnol. 2013, 97, 5413–5422. [Google Scholar] [CrossRef] [PubMed]
- Di Cagno, R.; De Angelis, M.; Limitone, A.; Minervini, F.; Carnevali, P.; Corsetti, A.; Gaenzle, M.; Ciati, R.; Gobbetti, M. Glucan and fructan production by sourdough Weissella cibaria and Lactobacillus plantarum. J. Agric. Food Chem. 2006, 54, 9873–9881. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.Y.; Lee, S.H.; Jeon, C.O. Kimchi microflora: History, current status, and perspectives for industrial kimchi production. Appl. Microbiol. Biotechnol. 2014, 98, 2385–2393. [Google Scholar] [CrossRef] [PubMed]
- Nuraida, L.; Wacher, M.C.; Owens, J.D. Microbiology of pozol, a Mexican maize dough. World J. Microbiol. Biotechnol. 1995, 11, 567–571. [Google Scholar] [CrossRef] [PubMed]
- Wolkers-Rooijackers, J.C.M.; Thomas, S.M.; Nout, M.J.R. Effects of sodium reduction scenarios on fermentation and quality of sauerkraut. LWT Food Sci. Technol. 2013, 54, 383–388. [Google Scholar] [CrossRef]
- Lynch, K.M.; Lucid, A.; Arendt, E.K.; Sleator, R.D.; Lucey, B.; Coffey, A. Genomics of Weissella cibaria with an examination of its metabolic traits. Microbiology 2015, 161, 914–930. [Google Scholar] [CrossRef] [PubMed]
- Chelo, I.M.; Zé-Zé, L.; Tenreiro, R. Genome diversity in the genera Fructobacillus, Leuconostoc and Weissella determined by physical and genetic mapping. Microbiology 2010, 156, 420–430. [Google Scholar] [CrossRef] [PubMed]
- Padonou, S.W.; Schillinger, U.; Nielsen, D.S.; Franz, C.M.A.P.; Hansen, M.; Hounhouigan, J.D.; Nago, M.C.; Jakobsen, M. Weissella beninensis sp. nov., a motile lactic acid bacterium from submerged cassava fermentations, and emended description of the genus Weissella. Int. J. Syst. Evol. Microbiol. 2010, 60, 2193–2198. [Google Scholar] [CrossRef] [PubMed]
- Vela, A.I.; Fernández, A.; de Quirós, Y.B.; Herráez, P.; Domínguez, L.; Fernández-Garayzábal, J.F. Weissella ceti sp. nov., isolated from beaked whales (Mesoplodon bidens). Int. J. Syst. Evol. Microbiol. 2011, 61, 2758–2762. [Google Scholar] [CrossRef] [PubMed]
- Fusco, V.; Quero, G.M.; Cho, G.-S.; Kabisch, J.; Meske, D.; Neve, H.; Bockelmann, W.; Franz, C.M.A.P. The genus Weissella: Taxonomy, ecology and biotechnological potential. Front. Microbiol. 2015, 6, 155. [Google Scholar] [CrossRef] [PubMed]
- Collins, M.D.; Samelis, J.; Metaxopoulos, J.; Wallbanks, S. Taxonomic studies on some Leuconostoc-like organisms from fermented sausages: Description of a new genus Weissella for the Leuconostoc paramesenteroides group of species. J. Appl. Bacteriol. 1993, 75, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Kandler, O.; Schillinger, U.; Weiss, N. Lactobacillus halotolerans sp.nov., nom.rev. and Lactobacillus minor sp.nov., nom.rev. Syst. Appl. Microbiol. 1983, 4, 280–285. [Google Scholar] [CrossRef]
- Björkroth, J.; Holzapfel, W. Genera Leuconostoc, Oenococcus and Weissella. In The Prokaryotes; Springer: New York, NY, USA, 2006; pp. 267–319. [Google Scholar]
- Dicks, L.M.T.; Dellaglio, F.; Collins, M.D. Proposal To Reclassify Leuconostoc oenos as Oenococcus oeni (corrig.) gen. nov., comb. nov. Int. J. Syst. Bacteriol. 1995, 45, 395–397. [Google Scholar] [CrossRef] [PubMed]
- Endo, A.; Okada, S. Reclassification of the genus Leuconostoc and proposals of Fructobacillus fructosus gen. nov., comb. nov., Fructobacillus durionis comb. nov.; Fructobacillus ficulneus comb. nov. and Fructobacillus pseudoficulneus comb. nov. Int. J. Syst. Evol. Microbiol. 2008, 58, 2195–2205. [Google Scholar] [CrossRef] [PubMed]
- Niven, C.F.; Evans, J.B. Lactobacillus viridescens nov. spec., a heterofermentative species that produces a green discoloration of cured meat pigments. J. Bacteriol. 1957, 73, 758–759. [Google Scholar] [PubMed]
- Nisiotou, A.; Dourou, D.; Filippousi, M.-E.; Banilas, G.; Tassou, C. Weissella uvarum sp. nov.; isolated from wine grapes. Int. J. Syst. Evol. Microbiol. 2014, 64, 3885–3890. [Google Scholar] [CrossRef] [PubMed]
- Snauwaert, I.; Papalexandratou, Z.; De Vuyst, L.; Vandamme, P. Characterization of strains of Weissella fabalis sp. nov. and Fructobacillus tropaeoli from spontaneous cocoa bean fermentations. Int. J. Syst. Evol. Microbiol. 2013, 63, 1709–1716. [Google Scholar] [CrossRef] [PubMed]
- De Bruyne, K.; Camu, N.; De Vuyst, L.; Vandamme, P. Weissella fabaria sp. nov.; from a Ghanaian cocoa fermentation. Int. J. Syst. Evol. Microbiol. 2010, 60, 1999–2005. [Google Scholar] [CrossRef] [PubMed]
- De Bruyne, K.; Camu, N.; Lefebvre, K.; De Vuyst, L.; Vandamme, P. Weissella ghanensis sp. nov.; isolated from a Ghanaian cocoa fermentation. Int. J. Syst. Evol. Microbiol. 2008, 58, 2721–2725. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-S.; Lee, S.-H.; Ahn, M.-J.; Ku, H.-J.; Hong, J.-S.; Jeon, C.O.; Lee, J.-H.; Lee, S.H.; Ryu, S.; Shin, H.; et al. Weissella jogaejeotgali sp. nov., isolated from jogae jeotgal, a traditional Korean fermented seafood. Int. J. Syst. Evol. Microbiol. 2015, 65, 4674–4681. [Google Scholar] [CrossRef] [PubMed]
- Tanasupawat, S.; Shida, O.; Okada, S.; Komagata, K. Lactobacillus acidipiscis sp. nov. and Weissella thailandensis sp. nov., isolated from fermented fish in Thailand. Int. J. Syst. Evol. Microbiol. 2000, 50, 1479–1485. [Google Scholar] [CrossRef] [PubMed]
- Fessard, A.; Kapoor, A.; Patche, J.; Assemat, S.; Hoarau, M.; Bourdon, E.; Bahorun, T.; Remize, F. Lactic Fermentation as an Efficient Tool to Enhance the Antioxidant Activity of Tropical Fruit Juices and Teas. Microorganisms 2017, 5, 23. [Google Scholar] [CrossRef] [PubMed]
- Garvie, E.I. The Growth Factor and Amino Acid Requirements of Species of the Genus Leuconostoc, including Leuconostoc paramesenteroides (sp.nov.) and Leuconostoc oenos. J. Gen. Microbiol. 1967, 48, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Praet, J.; Meeus, I.; Cnockaert, M.; Houf, K.; Smagghe, G.; Vandamme, P. Novel lactic acid bacteria isolated from the bumble bee gut: Convivina intestini gen. nov., sp. nov., Lactobacillus bombicola sp. nov., and Weissella bombi sp. nov. Antonie Leeuwenhoek 2015, 107, 1337–1349. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.J.; Shin, N.R.; Hyun, D.W.; Kim, P.S.; Kim, J.Y.; Kim, M.S.; Yun, J.H.; Bae, J.W. Weissella diestrammenae sp. nov.; isolated from the gut of a camel cricket (Diestrammena coreana). Int. J. Syst. Evol. Microbiol. 2013, 63, 2951–2956. [Google Scholar] [CrossRef] [PubMed]
- Holzapfel, W.H.; Van Wyk, E.P. Lactobacillus kandleri sp. nov., a new species of the subgenus betabacterium, with glycine in the peptidoglycan. Zentralblatt Bakteriol. Mikrobiol. Hyg. I. Abt. Orig. C Allg. Angew. Ökologische Mikrobiol. 1982, 3, 495–502. [Google Scholar] [CrossRef]
- Lee, J.; Lee, K.C.; Ahn, J.; Mheen, T.; Pyun, Y.; Park, Y. Weissella koreensis sp. nov.; isolated from kimchi. Int. J. Syst. Evol. Microbiol. 2002, 52, 1257–1261. [Google Scholar] [CrossRef] [PubMed]
- Björkroth, K.J.; Schillinger, U.; Geisen, R.; Weiss, N.; Hoste, B.; Holzapfel, W.H.; Korkeala, H.J.; Vandamme, P. Taxonomic study of Weissella confusa and description of Weissella cibaria sp. nov.; detected in food and clinical samples. Int. J. Syst. Evolut. Microbiol. 2002, 52, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Russo, P.; Beleggia, R.; Ferrer, S.; Pardo, I.; Spano, G. A polyphasic approach in order to identify dominant lactic acid bacteria during pasta manufacturing. LWT Food Sci. Technol. 2010, 43, 982–986. [Google Scholar] [CrossRef]
- Osimani, A.; Garofalo, C.; Aquilanti, L.; Milanović, V.; Clementi, F. Unpasteurised commercial boza as a source of microbial diversity. Int. J. Food Microbiol. 2015, 194, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Magnusson, J.; Jonsson, H.; Schnurer, J.; Roos, S. Weissella soli sp. nov., a lactic acid bacterium isolated from soil. Int. J. Syst. Evol. Microbiol. 2002, 52, 831–834. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.; Lee, D.; Yang, C.; Jeon, J.; Kim, J.; Han, H. Microbial population dynamics of kimchi, a fermented cabbage product. FEMS Microbiol. Lett. 2006, 257, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.H.; Lee, S.H.; Jung, J.Y.; Choi, E.J.; Jeon, C.O. Microbial succession and metabolite changes during long-term storage of Kimchi. J. Food Sci. 2013, 78, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Cho, Y.; Lee, Y.; Han, S.-K.; Kim, C.-G.; Choo, D.-W.; Kim, Y.-R.; Kim, H.-Y. A proteomic approach for rapid identification of Weissella species isolated from Korean fermented foods on MALDI-TOF MS supplemented with an in-house database. Int. J. Food Microbiol. 2017, 243, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Chun, J. Bacterial community structure in kimchi, a Korean fermented vegetable food, as revealed by 16S rRNA gene analysis. Int. J. Food Microbiol. 2005, 103, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Seo, H.N.; Hwang, T.S.; Lee, S.H.; Park, D.H. Characterization of exopolysaccharide (EPS) produced by Weissella cibaria SKkimchi3 isolated from kimchi. J. Microbiol. 2008, 46, 535–541. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Heo, G.Y.; Jun, W.L.; Oh, Y.J.; Park, J.A.; Park, Y.H.; Pyun, Y.R.; Jong, S.A. Analysis of kimchi microflora using denaturing gradient gel electrophoresis. Int. J. Food Microbiol. 2005, 102, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Lan, W.-T.T.; Chen, Y.-S.; Yanagida, F. Isolation and characterization of lactic acid bacteria from Yan-dong-gua (fermented wax gourd), a traditional fermented food in Taiwan. J. Biosci. Bioeng. 2009, 108, 484–487. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-S.; Wu, H.-C.; Wang, C.-M.; Lin, C.-C.; Chen, Y.-T.; Jhong, Y.-J.; Yanagida, F. Isolation and characterization of lactic acid bacteria from pobuzihi (fermented cummingcordia), a traditional fermented food in Taiwan. Folia Microbiol. 2013, 58, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.S.; Wu, H.; Lo, H.; Lin, W.; Hsu, W.; Lin, C.; Lin, P.; Yanagida, F. Isolation and characterisation of lactic acid bacteria from jiang-gua (fermented cucumbers), a traditional fermented food in Taiwan. J. Sci. Food Agric. 2012, 92, 2069–2075. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-S.; Liou, M.S.; Ji, S.H.; Yu, C.R.; Pan, S.F.; Yanagida, F. Isolation and characterization of lactic acid bacteria from Yan-tsai-shin (fermented broccoli stems), a traditional fermented food in Taiwan. J. Appl. Microbiol. 2013, 115, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-H.; Chen, Y.-S.; Yanagida, F. Isolation and characterisation of lactic acid bacteria from yan-jiang (fermented ginger), a traditional fermented food in Taiwan. J. Sci. Food Agric. 2011, 91, 1746–1750. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-S.; Wu, H.C.; Pan, S.F.; Lin, B.G.; Lin, Y.H.; Tung, W.C.; Li, Y.L.; Chiang, C.M.; Yanagida, F. Isolation and characterization of lactic acid bacteria from yan-taozih (pickled peaches) in Taiwan. Ann. Microbiol. 2013, 63, 607–614. [Google Scholar] [CrossRef]
- Chen, Y.-S.S.; Wu, H.C.; Yu, C.R.; Chen, Z.Y.; Lu, Y.C.; Yanagida, F. Isolation and characterization of lactic acid bacteria from xi-gua-mian (fermented watermelon), a traditional fermented food in Taiwan. Ital. J. Food Sci. 2016, 28, 9–14. [Google Scholar] [CrossRef]
- Chen, Y.-S.; Yanagida, F.; Hsu, J.S. Isolation and characterization of lactic acid bacteria from dochi (fermented black beans), a traditional fermented food in Taiwan. Lett. Appl. Microbiol. 2006, 43, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Anandharaj, M.; Sivasankari, B.; Santhanakaruppu, R.; Manimaran, M.; Rani, R.P.; Sivakumar, S. Determining the probiotic potential of cholesterol-reducing Lactobacillus and Weissella strains isolated from gherkins (fermented cucumber) and south Indian fermented koozh. Res. Microbiol. 2015, 166, 428–439. [Google Scholar] [CrossRef] [PubMed]
- Chao, S.-H.; Wu, R.-J.; Watanabe, K.; Tsai, Y.-C. Diversity of lactic acid bacteria in suan-tsai and fu-tsai, traditional fermented mustard products of Taiwan. Int. J. Food Microbiol. 2009, 135, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Chao, S.-H.; Tomii, Y.; Watanabe, K.; Tsai, Y.-C. Diversity of lactic acid bacteria in fermented brines used to make stinky tofu. Int. J. Food Microbiol. 2008, 123, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Miyashita, M.; Malimas, T.; Sugimoto, M.; Yoshino, M. 16S rRNA gene sequence analysis of lactic acid bacteria isolated from fermented foods in Thailand. Microb. Cult. Coll. 2012, 28, 1–9. [Google Scholar]
- Roh, S.W.; Kim, K.; Nam, Y.; Chang, H.; Park, E.; Bae, J.-W. Investigation of archaeal and bacterial diversity in fermented seafood using barcoded pyrosequencing. ISME J. 2009, 4, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.; Prajapati, J.B.; Holst, O.; Ljungh, A. Probiotic properties of exopolysaccharide producing lactic acid bacteria isolated from vegetables and traditional Indian fermented foods. Food Biosci. 2012, 5, 27–33. [Google Scholar] [CrossRef]
- Yuliana, N.; Dizon, E.I. Phenotypic Identification of Lactic Acid Bacteria Isolated from Tempoyak (Fermented Durian) Made in the Philippines. Int. J. Biol. 2011, 3, 145–152. [Google Scholar] [CrossRef]
- Lu, Z.; Pérez-Díaz, I.M.; Hayes, J.S.; Breidt, F. Bacteriophage ecology in a commercial cucumber fermentation. Appl. Environ. Microbiol. 2012, 78, 8571–8578. [Google Scholar] [CrossRef] [PubMed]
- Wouters, D.; Grosu-Tudor, S.; Zamfir, M.; De Vuyst, L. Bacterial community dynamics, lactic acid bacteria species diversity and metabolite kinetics of traditional Romanian vegetable fermentations. J. Sci. Food Agric. 2013, 93, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ji, Y.; Park, H.; Lee, J.; Park, S.; Yeo, S.; Shin, H.; Holzapfel, W.H. Selection of functional lactic acid bacteria as starter cultures for the fermentation of Korean leek (Allium tuberosum Rottler ex Sprengel.). Int. J. Food Microbiol. 2014, 191, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Wouters, D.; Bernaert, N.; Conjaerts, W.; Van Droogenbroeck, B.; de Loose, M.; De Vuyst, L. Species diversity, community dynamics, and metabolite kinetics of spontaneous leek fermentations. Food Microbiol. 2013, 33, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Breidt, F.; Plengvidhya, V.; Fleming, H.P. Bacteriophage ecology in commercial sauerkraut fermentations. Appl. Environ. Microbiol. 2003, 69, 3192–3202. [Google Scholar] [CrossRef] [PubMed]
- Plengvidhya, V.; Breidt, F.; Lu, Z.; Fleming, H.P. DNA fingerprinting of lactic acid bacteria in sauerkraut fermentations. Appl. Environ. Microbiol. 2007, 73, 7697–7702. [Google Scholar] [CrossRef] [PubMed]
- Ayeni, F.A.; Sánchez, B.; Adeniyi, B.A.; de Los Reyes-Gavilán, C.G.; Margolles, A.; Ruas-Madiedo, P. Evaluation of the functional potential of Weissella and Lactobacillus isolates obtained from Nigerian traditional fermented foods and cow’s intestine. Int. J. Food Microbiol. 2011, 147, 97–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akabanda, F.; Owusu-Kwarteng, J.; Tano-Debrah, K.; Glover, R.L.K.; Nielsen, D.S.; Jespersen, L. Taxonomic and molecular characterization of lactic acid bacteria and yeasts in nunu, a Ghanaian fermented milk product. Food Microbiol. 2013, 34, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Mathara, J.M.; Schillinger, U.; Kutima, P.M.; Mbugua, S.K.; Holzapfel, W.H. Isolation, identification and characterisation of the dominant microorganisms of kule naoto: The Maasai traditional fermented milk in Kenya. Int. J. Food Microbiol. 2004, 94, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Isono, Y.; Shingu, I.; Shimizu, S. Identification and Characteristics of Lactic Acid Bacteria Isolated from Masai Fermented Milk in Northern Tanzania. Biosci. Biotechnol. Biochem. 1994, 58, 660–664. [Google Scholar] [CrossRef]
- Kimura, M.; Danno, K.; Yasui, H. Immunomodulatory Function and Probiotic Properties of Lactic Acid Bacteria Isolated from Mongolian Fermented Milk. Bioscience 2006, 25, 147–155. [Google Scholar] [CrossRef]
- Rahman, N.; Xiaohong, C.; Meiqin, F.; Mingsheng, D. Characterization of the dominant microflora in naturally fermented camel milk Shubat. World J. Microbiol. Biotechnol. 2009, 25, 1941–1946. [Google Scholar] [CrossRef]
- Di Cagno, R.; Buchin, S.; de Candia, S.; De Angelis, M.; Fox, P.F.; Gobbetti, M. Characterization of Italian cheeses ripened under nonconventional conditions. J. Dairy Sci. 2007, 90, 2689–2704. [Google Scholar] [CrossRef] [PubMed]
- Morales, F.; Morales, J.I.; Hernández, C.H.; Hernández-Sánchez, H. Isolation and partial characterization of halotolerant lactic acid bacteria from two mexican cheeses. Appl. Biochem. Biotechnol. 2011, 164, 889–905. [Google Scholar] [CrossRef] [PubMed]
- Gerasi, E.; Litopoulou-Tzanetaki, E.; Tzanetakis, N. Microbiological study of Manura, a hard cheese made from raw ovine milk in the Greek island Sifnos. Int. J. Dairy Technol. 2003, 56, 117–122. [Google Scholar] [CrossRef]
- Baruzzi, F.; Matarante, A.; Morea, M.; Cocconcelli, P.S. Microbial community dynamics during the Scamorza Altamurana cheese natural fermentation. J. Dairy Sci. 2002, 85, 1390–1397. [Google Scholar] [CrossRef]
- Morea, M.; Baruzzi, F.; Cappa, F.; Cocconcelli, P.S. Molecular characterization of the Lactobacillus community in traditional processing of Mozzarella cheese. Int. J. Food Microbiol. 1998, 43, 53–60. [Google Scholar] [CrossRef]
- Williams, A.G.; Banks, J.M. Proteolytic and other hydrolytic enzyme activities in non-starter lactic acid bacteria (NSLAB) isolated from Cheddar cheese manufactured in the United Kingdom. Int. Dairy J. 1997, 7, 763–774. [Google Scholar] [CrossRef]
- Zamfir, M.; Vancanneyt, M.; Makras, L.; Vaningelgem, F.; Lefebvre, K.; Pot, B.; Swings, J.; De Vuyst, L. Biodiversity of lactic acid bacteria in Romanian dairy products. Syst. Appl. Microbiol. 2006, 29, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wang, W.H.; Menghe, B.L.; Jiri, M.T.; Wang, H.M.; Liu, W.J.; Bao, Q.H.; Lu, Q.; Zhang, J.C.; Wang, F.; et al. Diversity of lactic acid bacteria associated with traditional fermented dairy products in Mongolia. J. Dairy Sci. 2011, 94, 3229–3241. [Google Scholar] [CrossRef] [PubMed]
- Mugula, J.; Nnko, S.A.; Narvhus, J.; Sørhaug, T. Microbiological and fermentation characteristics of togwa, a Tanzanian fermented food. Int. J. Food Microbiol. 2003, 80, 187–199. [Google Scholar] [CrossRef]
- Djeni, N.T.; Bouatenin, K.M.J.-P.; Assohoun, N.M.C.; Toka, D.M.; Menan, E.H.; Dousset, X.; Dje, K.M. Biochemical and microbial characterization of cassava inocula from the three main attieke production zones in Côte d’Ivoire. Food Control 2015, 50, 133–140. [Google Scholar] [CrossRef]
- Kostinek, M.; Specht, I.; Edward, V.A.A.; Pinto, C.; Egounlety, M.; Sossa, C.; Mbugua, S.; Dortu, C.; Thonart, P.; Taljaard, L.; et al. Characterisation and biochemical properties of predominant lactic acid bacteria from fermenting cassava for selection as starter cultures. Int. J. Food Microbiol. 2007, 114, 342–351. [Google Scholar] [CrossRef] [PubMed]
- Kostinek, M.; Specht, I.; Edward, V.A.; Schillinger, U.; Hertel, C.; Holzapfel, W.H.; Franz, C.M.A.P. Diversity and technological properties of predominant lactic acid bacteria from fermented cassava used for the preparation of Gari, a traditional African food. Syst. Appl. Microbiol. 2005, 28, 527–540. [Google Scholar] [CrossRef] [PubMed]
- Padonou, W.S.; Nielsen, D.S.; Hounhouigan, J.D.; Thorsen, L.; Nago, M.C.; Jakobsen, M. The microbiota of Lafun, an African traditional cassava food product. Int. J. Food Microbiol. 2009, 133, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Kavitake, D.; Devi, P.B.; Singh, S.P.; Shetty, P.H. Characterization of a novel galactan produced by Weissella confusa KR780676 from an acidic fermented food. Int. J. Biol. Macromol. 2016, 86, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Bounaix, M.-S.; Robert, H.; Gabriel, V.; Morel, S.; Remaud-Siméon, M.; Gabriel, B.; Fontagné-Faucher, C. Characterization of dextran-producing Weissella strains isolated from sourdoughs and evidence of constitutive dextransucrase expression. FEMS Microbiol. Lett. 2010, 311, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Bounaix, M.S.; Gabriel, V.; Morel, S.; Robert, H.; Rabier, P.; Remaud-Siméon, M.; Gabriel, B.; Fontagné-Faucher, C. Biodiversity of exopolysaccharides produced from sucrose by sourdough lactic acid bacteria. J. Agric. Food Chem. 2009, 57, 10889–10897. [Google Scholar] [CrossRef] [PubMed]
- Moroni, A.V.; Arendt, E.K.; Dal Bello, F. Biodiversity of lactic acid bacteria and yeasts in spontaneously-fermented buckwheat and teff sourdoughs. Food Microbiol. 2011, 28, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Pontonio, E.; Nionelli, L.; Curiel, J.A.; Sadeghi, A.; Di Cagno, R.; Gobbetti, M.; Rizzello, C.G. Iranian wheat flours from rural and industrial mills: Exploitation of the chemical and technology features, and selection of autochthonous sourdough starters for making breads. Food Microbiol. 2015, 47, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Robert, H.; Gabriel, V.; Fontagné-Faucher, C. Biodiversity of lactic acid bacteria in French wheat sourdough as determined by molecular characterization using species-specific PCR. Int. J. Food Microbiol. 2009, 135, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Zotta, T.; Ricciardi, A.; Parente, E. Enzymatic activities of lactic acid bacteria isolated from Cornetto di Matera sourdoughs. Int. J. Food Microbiol. 2007, 115, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Ampe, F.; Ben Omar, N.; Moizan, C.; Wacher, C.; Guyot, J.P. Polyphasic study of the spatial distribution of microorganisms in Mexican pozol, a fermented maize dough, demonstrates the need for cultivation-independent methods to investigate traditional fermentations. Appl. Environ. Microbiol. 1999, 65, 5464–5473. [Google Scholar] [PubMed]
- Tohno, M.; Kitahara, M.; Inoue, H.; Uegaki, R.; Irisawa, T.; Ohkuma, M.; Tajima, K. Weissella oryzae sp. nov., isolated from fermented rice grains. Int. J. Syst. Evol. Microbiol. 2013, 63, 1417–1420. [Google Scholar] [CrossRef] [PubMed]
- Elizaquível, P.; Pérez-Cataluña, A.; Yépez, A.; Aristimuño, C.; Jiménez, E.; Cocconcelli, P.S.; Vignolo, G.; Aznar, R. Pyrosequencing vs. culture-dependent approaches to analyze lactic acid bacteria associated to chicha, a traditional maize-based fermented beverage from Northwestern Argentina. Int. J. Food Microbiol. 2015, 198, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Oguntoyinbo, F.A.; Tourlomousis, P.; Gasson, M.J.; Narbad, A. Analysis of bacterial communities of traditional fermented West African cereal foods using culture independent methods. Int. J. Food Microbiol. 2011, 145, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Owusu-Kwarteng, J.; Akabanda, F.; Nielsen, D.S.; Tano-Debrah, K.; Glover, R.L.K.; Jespersen, L. Identification of lactic acid bacteria isolated during traditional fura processing in Ghana. Food Microbiol. 2012, 32, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Abegaz, K. Isolation, characterization and identification of lactic acid bacteria involved in traditional fermentation of borde, an Ethiopian cereal beverage. Afr. J. Biotechnol. 2007, 6, 1469–1478. [Google Scholar]
- Camu, N.; De Winter, T.; Verbrugghe, K.; Cleenwerck, I.; Vandamme, P.; Takrama, J.S.; Vancanneyt, M.; De Vuyst, L. Dynamics and biodiversity of populations of lactic acid bacteria and acetic acid bacteria involved in spontaneous heap fermentation of cocoa beans in Ghana. Appl. Environ. Microbiol. 2007, 73, 1809–1824. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.-J.; Nam, Y.-D.; Roh, S.W.; Bae, J.-W. Unexpected convergence of fungal and bacterial communities during fermentation of traditional Korean alcoholic beverages inoculated with various natural starters. Food Microbiol. 2012, 30, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Pringsulaka, O.; Patarasinpaiboon, N.; Suwannasai, N.; Atthakor, W.; Rangsiruji, A. Isolation and characterisation of a novel Podoviridae-phage infecting Weissella cibaria N 22 from Nham, a Thai fermented pork sausage. Food Microbiol. 2011, 28, 518–525. [Google Scholar] [CrossRef] [PubMed]
- Thongsanit, J.; Tanikawa, M.; Yano, S.; Tachiki, T.; Wakayama, M. Identification of glutaminase-producing lactic acid bacteria isolated from Nham, a traditional Thai fermented food and characterisation of glutaminase activity of isolated Weissella cibaria. Ann. Microbiol. 2009, 59, 715–720. [Google Scholar] [CrossRef]
- Wongsuphachat, W.; H-Kittikun, A.; Maneerat, S. Optimization of exopolysaccharides production by Weissella confusa NH 02 isolated from thai fermented sausages. Songklanakarin J. Sci. Technol. 2010, 32, 27–35. [Google Scholar]
- Cocolin, L.; Dolci, P.; Rantsiou, K.; Urso, R.; Cantoni, C.; Comi, G. Lactic acid bacteria ecology of three traditional fermented sausages produced in the North of Italy as determined by molecular methods. Meat Sci. 2009, 82, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Urso, R.; Comi, G.; Cocolin, L. Ecology of lactic acid bacteria in Italian fermented sausages: Isolation, identification and molecular characterization. Syst. Appl. Microbiol. 2006, 29, 671–680. [Google Scholar] [CrossRef] [PubMed]
- Papamanoli, E.; Tzanetakis, N.; Litopoulou-Tzanetaki, E.; Kotzekidou, P. Characterization of lactic acid bacteria isolated from a Greek dry-fermented sausage in respect of their technological and probiotic properties. Meat Sci. 2003, 65, 859–867. [Google Scholar] [CrossRef]
- Rantsiou, K.; Drosinos, E.H.; Gialitaki, M.; Urso, R.; Krommer, J.; Gasparik-Reichardt, J.; Tóth, S.; Metaxopoulos, I.; Comi, G.; Cocolin, L. Molecular characterization of Lactobacillus species isolated from naturally fermented sausages produced in Greece, Hungary and Italy. Food Microbiol. 2005, 22, 19–28. [Google Scholar] [CrossRef]
- Albano, H.; van Reenen, C.A.; Todorov, S.D.; Cruz, D.; Fraga, L.; Hogg, T.; Dicks, L.M.T.; Teixeira, P. Phenotypic and genetic heterogeneity of lactic acid bacteria isolated from “Alheira”, a traditional fermented sausage produced in Portugal. Meat Sci. 2009, 82, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Yüceer, Ö.; Tuncer, B.O. Determination of antibiotic resistance and biogenic amine production of lactic acid bacteria isolated from fermented Turkish sausage (sucuk). J. Food Saf. 2015, 35, 276–285. [Google Scholar] [CrossRef]
- Pereira, C.I.; San Romão, M.V.; Lolkema, J.S.; Crespo, M.T.B. Weissella halotolerans W22 combines arginine deiminase and ornithine decarboxylation pathways and converts arginine to putrescine. J. Appl. Microbiol. 2009, 107, 1894–1902. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Ji, H.; Wang, Q.; Li, B.; Li, K.; Xu, C.; Jiang, C. The effects of starter cultures and plant extracts on the biogenic amine accumulation in traditional Chinese smoked horsemeat sausages. Food Control 2015, 50, 869–875. [Google Scholar] [CrossRef]
- Samelis, J.; Maurogenakis, F.; Metaxopoulos, J. Characterisation of lactic acid bacteria isolated from naturally fermented Greek dry salami. Int. J. Food Microbiol. 1994, 23, 179–196. [Google Scholar] [CrossRef]
- Chen, C.; Chen, X.; Jiang, M.; Rui, X.; Li, W.; Dong, M. A newly discovered bacteriocin from Weissella cibaria D1501 associated with Chinese Dong fermented meat (Nanx Wudl). Food Control 2014, 42, 116–124. [Google Scholar] [CrossRef]
- Kopermsub, P.; Yunchalard, S. Identification of lactic acid bacteria associated with the production of plaa-som, a traditional fermented fish product of Thailand. Int. J. Food Microbiol. 2010, 138, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Paludan-Müller, C.; Madsen, M.; Sophanodora, P.; Gram, L.; Møller, P.L. Fermentation and microflora of plaa-som, a Thai fermented fish product prepared with different salt concentrations. Int. J. Food Microbiol. 2002, 73, 61–70. [Google Scholar] [CrossRef]
- Srionnual, S.; Yanagida, F.; Lin, L.H.; Hsiao, K.N.; Chen, Y.S. Weissellicin 110, a newly discovered bacteriocin from Weissella cibaria 110, isolated from plaa-som, a fermented fish product from Thailand. Appl. Environ. Microbiol. 2007, 73, 2247–2250. [Google Scholar] [CrossRef] [PubMed]
- Thapa, N.; Pal, J.; Prakash, J. Phenotypic identification and technological properties of lactic acid bacteria isolated from traditionally processed fish products of the Eastern Himalayas. Int. J. Food Microbiol. 2006, 107, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.; Falck, P.; Shah, N.; Immerzeel, P.; Adlercreutz, P.; Stålbrand, H.; Prajapati, J.B.; Holst, O.; Nordberg Karlsson, E. Evidence for xylooligosaccharide utilization in Weissella strains isolated from Indian fermented foods and vegetables. FEMS Microbiol. Lett. 2013, 346, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Swain, M.R.; Anandharaj, M.; Ray, R.C.; Praveen Rani, R. Fermented fruits and vegetables of Asia: A potential source of probiotics. Biotechnol. Res. Int. 2014, 2014, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Holzapfel, W. Use of starter cultures in fermentation on a household scale. Food Control 1997, 8, 241–258. [Google Scholar] [CrossRef]
- Marsh, A.J.; Hill, C.; Ross, R.P.; Cotter, P.D. Fermented beverages with health-promoting potential: Past and future perspectives. Trends Food Sci. Technol. 2014, 38, 113–124. [Google Scholar] [CrossRef] [Green Version]
- Holzapfel, W.H. Appropriate starter culture technologies for small-scale fermentation in developing countries. Int. J. Food Microbiol. 2002, 75, 197–212. [Google Scholar] [CrossRef]
- Ammor, M.S.; Mayo, B. Selection criteria for lactic acid bacteria to be used as functional starter cultures in dry sausage production: An update. Meat Sci. 2007, 76, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Iyer, R.; Tomar, S.K.; Uma Maheswari, T.; Singh, R. Streptococcus thermophilus strains: Multifunctional lactic acid bacteria. Int. Dairy J. 2010, 20, 133–141. [Google Scholar] [CrossRef]
- Ogier, J.C.; Casalta, E.; Farrokh, C.; Saïhi, A. Safety assessment of dairy microorganisms: The Leuconostoc genus. Int. J. Food Microbiol. 2008, 126, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Altuntas, E.G. Bacteriocins: A natural way to combat with pathogens. In Microbial Pathogens and Strategies for Combating Them: Science, Technology and Education; Méndez-Vilas, A., Ed.; Formatex: Badajoz, Spain, 2013; pp. 1007–1015. [Google Scholar]
- Rattanachaikunsopon, P.; Phumkhachorn, P. Lactic acid bacteria: Their antimicrobial compounds and their uses in food production. Ann. Biol. Res. 2010, 1, 218–228. [Google Scholar]
- Varsha, K.K.; Nampoothiri, K.M. Appraisal of lactic acid bacteria as protective cultures. Food Control 2016, 69, 61–64. [Google Scholar] [CrossRef]
- Hati, S.; Mandal, S.; Prajapati, J. Novel Starters for Value Added Fermented Dairy Products. Curr. Res. Nutr. Food Sci. J. 2013, 1, 83–91. [Google Scholar] [CrossRef]
- Hammes, W.P.; Knauf, H.J. Starters in the Processing of Meat-Products. Meat Sci. 1994, 36, 155–168. [Google Scholar] [CrossRef]
- Zannini, E.; Waters, D.M.; Coffey, A.; Arendt, E.K. Production, properties, and industrial food application of lactic acid bacteria-derived exopolysaccharides. Appl. Microbiol. Biotechnol. 2016, 100, 1121–1135. [Google Scholar] [CrossRef] [PubMed]
- Di Cagno, R.; Surico, R.F.; Siragusa, S.; De Angelis, M.; Paradiso, A.; Minervini, F.; De Gara, L.; Gobbetti, M. Selection and use of autochthonous mixed starter for lactic acid fermentation of carrots, French beans or marrows. Int. J. Food Microbiol. 2008, 127, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Dueñas, M.; Fernández, D.; Hernández, T.; Estrella, I.; Muñoz, R. Bioactive phenolic compounds of cowpeas (Vigna sinensis L). Modifications by fermentation with natural microflora and with Lactobacillus plantarum ATCC 14917. J. Sci. Food Agric. 2005, 85, 297–304. [Google Scholar] [CrossRef]
- Filannino, P.; Azzi, L.; Cavoski, I.; Vincentini, O.; Rizzello, C.G.; Gobbetti, M.; Di Cagno, R. Exploitation of the health-promoting and sensory properties of organic pomegranate (Punica granatum L.) juice through lactic acid fermentation. Int. J. Food Microbiol. 2013, 163, 184–192. [Google Scholar] [CrossRef] [PubMed]
- Marazza, J.A.; Nazareno, M.A.; Savoy de Giori, G.; Garro, M.S. Enhancement of the antioxidant capacity of soymilk by fermentation with Lactobacillus rhamnosus. J. Funct. Foods 2012, 4, 594–601. [Google Scholar] [CrossRef]
- Hur, S.J.; Lee, S.Y.; Kim, Y.-C.; Choi, I.; Kim, G.-B. Effect of fermentation on the antioxidant activity in plant-based foods. Food Chem. 2014, 160, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Shalaby, A.R. Significance of biogenic amines to food safety and human health. Food Res. Int. 1997, 29, 675–690. [Google Scholar] [CrossRef]
- Hansen, E.B. Commercial bacterial starter cultures for fermented foods of the future. Int. J. Food Microbiol. 2002, 78, 119–131. [Google Scholar] [CrossRef]
- Ahmed, F.E. Genetically modified probiotics in foods. Trends Biotechnol. 2003, 21, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Joerger, M.C.; Klaenhammer, T.R. Cloning, expression, and nucleotide sequence of the Lactobacillus helveticus 481 gene encoding the bacteriocin helveticin J. J. Bacteriol. 1990, 172, 6339–6347. [Google Scholar] [CrossRef] [PubMed]
- Joutsjoki, V.; Luoma, S.; Tamminen, M.; Kilpi, M.; Johansen, E.; Palva, A. Recombinant Lactococcus starters as a potential source of additional peptidolytic activity in cheese ripening. J. Appl. Microbiol. 2002, 92, 1159–1166. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, H.; Pronk, J.T.; Teusink, B. Evolutionary engineering to enhance starter culture performance in food fermentations. Curr. Opin. Biotechnol. 2015, 32, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Cakar, Z.P.; Turanli-Yildiz, B.; Alkim, C.; Yilmaz, U. Evolutionary engineering of Saccharomyces cerevisiae for improved industrially important properties. FEMS Yeast Res. 2012, 12, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Cakar, Z.P.; Seker, U.O.S.; Tamerler, C.; Sonderegger, M.; Sauer, U. Evolutionary engineering of multiple-stress resistant Saccharomyces cerevisiae. FEMS Yeast Res. 2005, 5, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, H.; Starrenburg, M.J.C.; Molenaar, D.; Kleerebezem, M.; Hylckama Vlieg, J.E.T. Microbial domestication signatures of Lactococcus lactis can be reproduced by experimental evolution. Genome Res. 2012, 22, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Kim, Y.-W.; Hwang, I.; Kim, J.; Yoon, S. Evaluation of Leuconostoc citreum HO12 and Weissella koreensis HO20 isolated from kimchi as a starter culture for whole wheat sourdough. Food Chem. 2012, 134, 2208–2216. [Google Scholar] [CrossRef] [PubMed]
- Robert, H.; Gabriel, V.; Lefebvre, D.; Rabier, P.; Vayssier, Y.; Fontagné-Faucher, C. Study of the behaviour of Lactobacillus plantarum and Leuconostoc starters during a complete wheat sourdough breadmaking process. LWT Food Sci. Technol. 2006, 39, 256–265. [Google Scholar] [CrossRef]
- Wolter, A.; Hager, A.-S.; Zannini, E.; Galle, S.; Gänzle, M.G.; Waters, D.M.; Arendt, E.K. Evaluation of exopolysaccharide producing Weissella cibaria MG1 strain for the production of sourdough from various flours. Food Microbiol. 2014, 37, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Di Cagno, R.; Surico, R.F.; Minervini, G.; De Angelis, M.; Rizzello, C.G.; Gobbetti, M. Use of autochthonous starters to ferment red and yellow peppers (Capsicum annum L.) to be stored at room temperature. Int. J. Food Microbiol. 2009, 130, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Galle, S.; Schwab, C.; Arendt, E.; Gänzle, M. Exopolysaccharide-forming Weissella strains as starter cultures for sorghum and wheat sourdoughs. J. Agric. Food Chem. 2010, 58, 5834–5841. [Google Scholar] [CrossRef] [PubMed]
- Malang, S.K.; Maina, N.H.; Schwab, C.; Tenkanen, M.; Lacroix, C. Characterization of exopolysaccharide and ropy capsular polysaccharide formation by Weissella. Food Microbiol. 2015, 46, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Zannini, E.; Mauch, A.; Galle, S.; Gänzle, M.; Coffey, A.; Arendt, E.K.; Taylor, J.P.; Waters, D.M. Barley malt wort fermentation by exopolysaccharide-forming Weissella cibaria MG1 for the production of a novel beverage. J. Appl. Microbiol. 2013, 115, 1379–1387. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, R.Z.; Siddiqui, K.; Arman, M.; Ahmed, N. Characterization of high molecular weight dextran produced by Weissella cibaria CMGDEX3. Carbohydr. Polym. 2012, 90, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Rao, T.J.M.; Goyal, A. A novel high dextran yielding Weissella cibaria JAG8 for cereal food application. Int. J. Food Sci. Nutr. 2013, 64, 346–354. [Google Scholar] [CrossRef] [PubMed]
- Tingirikari, J.M.R.; Kothari, D.; Shukla, R.; Goyal, A. Structural and biocompatibility properties of dextran from Weissella cibaria JAG8 as food additive. Int. J. Food Sci. Nutr. 2014, 65, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Maina, N.H.; Tenkanen, M.; Maaheimo, H.; Juvonen, R.; Virkki, L. NMR spectroscopic analysis of exopolysaccharides produced by Leuconostoc citreum and Weissella confusa. Carbohydr. Res. 2008, 343, 1446–1455. [Google Scholar] [CrossRef] [PubMed]
- Juvonen, R.; Honkapää, K.; Maina, N.H.; Shi, Q.; Viljanen, K.; Maaheimo, H.; Virkki, L.; Tenkanen, M.; Lantto, R. The impact of fermentation with exopolysaccharide producing lactic acid bacteria on rheological, chemical and sensory properties of pureed carrots (Daucus carota L.). Int. J. Food Microbiol. 2015, 207, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Nam, H.; Ha, M.; Bae, O.; Lee, Y. Effect of Weissella confusa strain PL9001 on the adherence and growth of Helicobacter pylori. Appl. Environ. Microbiol. 2002, 68, 4642–4645. [Google Scholar] [CrossRef] [PubMed]
- Leong, K.; Chen, Y.; Lin, Y.; Pan, S.; Yu, B.; Wu, H.; Yanagida, F. Weissellicin L, a novel bacteriocin from sian-sianzih-isolated Weissella hellenica 4–7. J. Appl. Microbiol. 2013, 115, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Masuda, Y.; Zendo, T.; Sawa, N.; Perez, R.H.; Nakayama, J.; Sonomoto, K. Characterization and identification of weissellicin Y and weissellicin M, novel bacteriocins produced by Weissella cibaria QU 13. J. Appl. Microbiol. 2012, 112, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Papagianni, M.; Papamichael, E.M. Purification, amino acid sequence and characterization of the class IIa bacteriocin weissellin A, produced by Weissella paramesenteroides DX. Bioresour. Technol. 2011, 102, 6730–6734. [Google Scholar] [CrossRef] [PubMed]
- Woraprayote, W.; Pumpuang, L.; Tosukhowong, A.; Roytrakul, S.; Perez, R.H.; Zendo, T.; Sonomoto, K.; Benjakul, S.; Visessanguan, W. Two putatively novel bacteriocins active against Gram-negative food borne pathogens produced by Weissella cibaria BCC 7293. Food Control 2015, 55, 176–184. [Google Scholar] [CrossRef]
- Malik, A.; Sumayyah, S.; Yeh, C.W.; Heng, N.C.K. Identification and sequence analysis of pWcMBF8-1, a bacteriocin-encoding plasmid from the lactic acid bacterium Weissella confusa. FEMS Microbiol. Lett. 2016, 363, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Parvez, S.; Malik, K.A.; Ah Kang, S.; Kim, H.-Y. Probiotics and their fermented food products are beneficial for health. J. Appl. Microbiol. 2006, 100, 1171–1185. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.W.; Park, J.Y.; Jeong, H.R.; Heo, H.J.; Han, N.S.; Kim, J.H. Probiotic properties of Weissella strains isolated from human faeces. Anaerobe 2012, 18, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Fritsch, C.; Jänsch, A.; Ehrmann, M.A.; Toelstede, S.; Vogel, R.F. Characterization of Cinnamoyl Esterases from Different Lactobacilli and Bifidobacteria. Curr. Microbiol. 2017, 74, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, N.; Esteban-Torres, M.; Mancheño, J.M.; de Las Rivas, B.; Muñoz, R. Tannin degradation by a novel tannase enzyme present in some Lactobacillus plantarum strains. Appl. Environ. Microbiol. 2014, 80, 2991–2997. [Google Scholar] [CrossRef] [PubMed]
- Pyo, Y.-H.; Lee, T.-C.; Lee, Y.-C. Enrichment of bioactive isoflavones in soymilk fermented with β-glucosidase-producing lactic acid bacteria. Food Res. Int. 2005, 38, 551–559. [Google Scholar] [CrossRef]
- Rodriguez, H.; Curiel, J.A.; Landete, J.M.; de las Rivas, B.; Lopez de Felipe, F.; Gomez-Cordoves, C.; Mancheno, J.M.; Munoz, R. Food phenolics and lactic acid bacteria. Int. J. Food Microbiol. 2009, 132, 79–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez, H.; Landete, J.M.; de las Rivas, B.; Muñoz, R. Metabolism of food phenolic acids by Lactobacillus plantarum CECT 748T. Food Chem. 2008, 107, 1393–1398. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.W.; Han, N.S.; Kim, J.H. Purification and characterization of beta-glucosidase from Weissella cibaria 37. J. Microbiol. Biotechnol. 2012, 22, 1705–1713. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Kim, G.M.; Lee, K.W.; Choi, I.D.; Kwon, G.H.; Park, J.Y.; Jeong, S.J.; Kim, J.S.; Kim, J.H. Conversion of isoflavone glucosides to aglycones in soymilk by fermentation with lactic acid bacteria. J. Food Sci. 2007, 72, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Kim, J.S.; Kim, J.H. Enrichment of isoflavone aglycones in soymilk by fermentation with single and mixed cultures of Streptococcus infantarius 12 and Weissella sp. 4. Food Chem. 2008, 109, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Filannino, P.; Gobbetti, M.; De Angelis, M.; Di Cagno, R. Hydroxycinnamic acids used as external acceptors of electrons: An energetic advantage for strictly heterofermentative lactic acid bacteria. Appl. Environ. Microbiol. 2014, 80, 7574–7582. [Google Scholar] [CrossRef] [PubMed]
- Park, J.A.; Tirupathi Pichiah, P.B.; Yu, J.J.; Oh, S.H.; Daily, J.W.; Cha, Y.S. Anti-obesity effect of kimchi fermented with Weissella koreensis OK1-6 as starter in high-fat diet-induced obese C57BL/6J mice. J. Appl. Microbiol. 2012, 113, 1507–1516. [Google Scholar] [CrossRef] [PubMed]
- Spano, G.; Russo, P.; Lonvaud-Funel, A.; Lucas, P.; Alexandre, H.; Grandvalet, C.; Coton, E.; Coton, M.; Barnavon, L.; Bach, B.; et al. Biogenic amines in fermented foods. Eur. J. Clin. Nutr. 2010, 64, S95–S100. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-atienza, E.; Landeta, G.; De, B.; Gómez-sala, B.; Muñoz, R.; Hernández, P.E.; Cintas, L.M.; Herranz, C. Phenotypic and genetic evaluations of biogenic amine production by lactic acid bacteria isolated from fish and fish products. Int. J. Food Microbiol. 2011, 146, 212–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, D.-W.; Lee, J.-H. Antibiotic resistance, hemolysis and biogenic amine production assessments of Leuconostoc and Weissella isolates for kimchi starter development. LWT Food Sci. Technol. 2015, 64, 1078–1084. [Google Scholar] [CrossRef]
- Arena, M.E.; Manca De Nadra, M.C. Biogenic amine production by Lactobacillus. J. Appl. Microbiol. 2001, 90, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Costantini, A.; Pietroniro, R.; Doria, F.; Pessione, E.; Garcia-Moruno, E. Putrescine production from different amino acid precursors by lactic acid bacteria from wine and cider. Int. J. Food Microbiol. 2013, 165, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Dapkevicius, M.L.N.E.; Nout, M.J.R.; Rombouts, F.M.; Houben, J.H.; Wymenga, W. Biogenic amine formation and degradation by potential fish silage starter microorganisms. Int. J. Food Microbiol. 2000, 57, 107–114. [Google Scholar] [CrossRef]
- Mathur, S.; Singh, R. Antibiotic resistance in food lactic acid bacteria—A review. Int. J. Food Microbiol. 2005, 105, 281–295. [Google Scholar] [CrossRef] [PubMed]
- D’Aimmo, M.R.; Modesto, M.; Biavati, B. Antibiotic resistance of lactic acid bacteria and Bifidobacterium spp. isolated from dairy and pharmaceutical products. Int. J. Food Microbiol. 2007, 115, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Tomar, S.K.; Goswami, P.; Sangwan, V.; Singh, R. Antibiotic resistance among commercially available probiotics. Food Res. Int. 2014, 57, 176–195. [Google Scholar] [CrossRef]
- Jin, L.; Tao, L.; Pavlova, S.I.; So, J.S.; Kiwanuka, N.; Namukwaya, Z.; Saberbein, B.A.; Wawer, M. Species diversity and relative abundance of vaginal lactic acid bacteria from women in Uganda and Korea. J. Appl. Microbiol. 2007, 102, 1107–1115. [Google Scholar] [CrossRef] [PubMed]
- Silvester, M.E.; Dicks, L.M.T. Identification of lactic acid bacteria isolated from human vaginal secretions. Antonie Leeuwenhoek 2003, 83, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y. Characterization of Weissella kimchii PL9023 as a potential probiotic for women. FEMS Microbiol. Lett. 2005, 250, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Gomathi, S.; Sasikumar, P.; Anbazhagan, K.; Sasikumar, S.; Kavitha, M.; Selvi, M.S.; Selvam, G.S. Screening of indigenous oxalate degrading lactic acid bacteria from human faeces and south Indian fermented foods: Assessment of probiotic potential. Sci. World J. 2014, 2014, 648059. [Google Scholar] [CrossRef] [PubMed]
- Nistal, E.; Caminero, A.; Vivas, S.; Ruiz De Morales, J.M.; Sáenz De Miera, L.E.; Rodríguez-Aparicio, L.B.; Casqueiro, J. Differences in faecal bacteria populations and faecal bacteria metabolism in healthy adults and celiac disease patients. Biochimie 2012, 94, 1724–1729. [Google Scholar] [CrossRef] [PubMed]
- Rubio, R.; Jofré, A.; Martín, B.; Aymerich, T.; Garriga, M. Characterization of lactic acid bacteria isolated from infant faeces as potential probiotic starter cultures for fermented sausages. Food Microbiol. 2014, 38, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Walter, J.; Hertel, C.; Tannock, G.W.; Lis, C.M.; Munro, K.; Hammes, W.P. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella Species in Human Feces by Using Group-Specific PCR Primers and Denaturing Gradient Gel Electrophoresis. Appl. Environ. Microbiol. 2001, 67, 2578–2585. [Google Scholar] [CrossRef] [PubMed]
- Costa, F.A.A.; Leal, C.A.G.; Schuenker, N.D.; Leite, R.C.; Figueiredo, H.C.P. Characterization of Weissella ceti infections in Brazilian rainbow trout, Oncorhynchus mykiss (Walbaum), farms and development of an oil-adjuvanted vaccine. J. Fish Dis. 2015, 38, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Snyder, A.K.; Hinshaw, J.M.; Welch, T.J. Diagnostic tools for rapid detection and quantification of Weissella ceti NC36 infections in rainbow trout. Lett. Appl. Microbiol. 2015, 60, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Abriouel, H.; Lerma, L.L.; Casado Muñoz, M.D.C.; Montoro, B.P.; Kabisch, J.; Pichner, R.; Cho, G.-S.; Neve, H.; Fusco, V.; Franz, C.M.A.P.; et al. The controversial nature of the Weissella genus: Technological and functional aspects versus whole genome analysis-based pathogenic potential for their application in food and health. Front. Microbiol. 2015, 6, 1197. [Google Scholar] [CrossRef] [PubMed]
- Fairfax, M.R.; Lephart, P.R.; Salimnia, H. Weissella confusa: Problems with identification of an opportunistic pathogen that has been found in fermented foods and proposed as a probiotic. Front. Microbiol. 2014, 5, 254. [Google Scholar] [CrossRef] [PubMed]
- Flaherty, J.D.; Levett, P.N.; Dewhirst, F.E.; Troe, T.E.; Warren, J.R.; Johnson, S. Fatal case of endocarditis due to Weissella confusa. J. Clin. Microbiol. 2003, 41, 2237–2239. [Google Scholar] [CrossRef] [PubMed]
- Harlan, N.P.; Kempker, R.R.; Parekh, S.M.; Burd, E.M.; Kuhar, D.T. Weissella confusa bacteremia in a liver transplant patient with hepatic artery thrombosis. Transpl. Infect. Dis. 2011, 13, 290–293. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Augustine, D.; Sudhindran, S.; Kurian, A.M.; Dinesh, K.R.; Karim, S.; Philip, R. Weissella confusa: A rare cause of vancomycinresistant Gram-positive bacteraemia. J. Med. Microbiol. 2011, 60, 1539–1541. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.R.; Huang, Y.T.; Liao, C.H.; Lai, C.C.; Lee, P.I.; Hsueh, P.R. Bacteraemia caused by Weissella confusa at a university hospital in Taiwan, 1997–2007. Clin. Microbiol. Infect. 2011, 17, 1226–1231. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Cho, S.M.; Kim, M.; Ko, Y.G.; Yong, D.; Lee, K. Weissella confusa bacteremia in an immune-competent patient with underlying intramural hematomas of the aorta. Ann. Lab. Med. 2013, 33, 459–462. [Google Scholar] [CrossRef] [PubMed]
- Medford, R.; Patel, S.N.; Evans, G.A. A confusing case—Weissella confusa prosthetic joint infection: A case report and review of the literature. Can. J. Infect. Dis. Med. Microbiol. 2014, 25, 173–175. [Google Scholar] [CrossRef] [PubMed]
- Olano, A.; Chua, J.; Schroeder, S.; Minari, A.; Salvia, M.L.A.; Hall, G. Weissella confusa (Basonym: Lactobacillus confusus) Bacteremia: A Case Report. J. Clin. Microbiol. 2001, 39, 1604–1607. [Google Scholar] [CrossRef] [PubMed]
- Salimnia, H.; Alangaden, G.J.; Bharadwaj, R.; Painter, T.M.; Chandrasekar, P.H.; Fairfax, M.R. Weissella confusa: An unexpected cause of vancomycin-resistant gram-positive bacteremia in immunocompromised hosts. Transpl. Infect. Dis. 2011, 13, 294–298. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.H.; Kim, D.I.L.; Kim, H.R.; Kim, D.S.; Kook, J.K.; Lee, J.N. Severe infective endocarditis of native valves caused by Weissella confusa detected incidentally on echocardiography. J. Infect. 2007, 54, 149–151. [Google Scholar] [CrossRef] [PubMed]
- Svec, P.; Sevcíková, A.; Sedlácek, I.; Bednárová, J.; Snauwaert, C.; Lefebvre, K.; Vandamme, P.; Vancanneyt, M. Identification of lactic acid bacteria isolated from human blood cultures. FEMS Immunol. Med. Microbiol. 2007, 49, 192–196. [Google Scholar] [CrossRef]
- Kamboj, K.; Vasquez, A.; Balada-Llasat, J.M. Identification and significance of Weissella species infections. Front. Microbiol. 2015, 6, 1204. [Google Scholar] [CrossRef] [PubMed]
- Cannon, J.P.; Lee, T.A.; Bolanos, J.T.; Danziger, L.H. Pathogenic relevance of Lactobacillus: A retrospective review of over 200 cases. Eur. J. Clin. Microbiol. Infect. Dis. 2005, 24, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Franko, B.; Fournier, P.; Jouve, T.; Malvezzi, P.; Pelloux, I.; Brion, J.P.; Pavese, P. Lactobacillus bacteremia: Pathogen or prognostic marker? Med. Mal. Infect. 2017, 47, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Garneau, J.E.; Moineau, S. Bacteriophages of lactic acid bacteria and their impact on milk fermentations. Microb. Cell Factories 2011, 10, S20. [Google Scholar] [CrossRef] [PubMed]
- Kot, W.; Neve, H.; Heller, K.J.; Vogensen, F.K. Bacteriophages of Leuconostoc, Oenococcus, and Weissella. Front. Microbiol. 2014, 5, 186. [Google Scholar] [CrossRef] [PubMed]
- Kleppen, H.P.; Holo, H.; Jeon, S.R.; Nes, I.F.; Yoon, S.S. Novel Podoviridae family bacteriophage infecting Weissella cibaria isolated from kimchi. Appl. Environ. Microbiol. 2012, 78, 7299–7308. [Google Scholar] [CrossRef] [PubMed]
- Mahony, J.; Ainsworth, S.; Stockdale, S.; van Sinderen, D. Phages of lactic acid bacteria: The role of genetics in understanding phage-host interactions and their co-evolutionary processes. Virology 2012, 434, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Klaenhammer, T.R.; Kullen, M.J. Selection and design of probiotics. Int. J. Food Microbiol. 1999, 50, 45–57. [Google Scholar] [CrossRef]
- Bourdichon, F.; Casaregola, S.; Farrokh, C.; Frisvad, J.C.; Gerds, M.L.; Hammes, W.P.; Harnett, J.; Huys, G.; Laulund, S.; Ouwehand, A.; et al. Food fermentations: Microorganisms with technological beneficial use. Int. J. Food Microbiol. 2012, 154, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Bevilacqua, A.; Corbo, M.R.; Sinigaglia, M. Selection of Yeasts as Starter Cultures for Table Olives: A Step-by-Step Procedure. Front. Microbiol. 2012, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
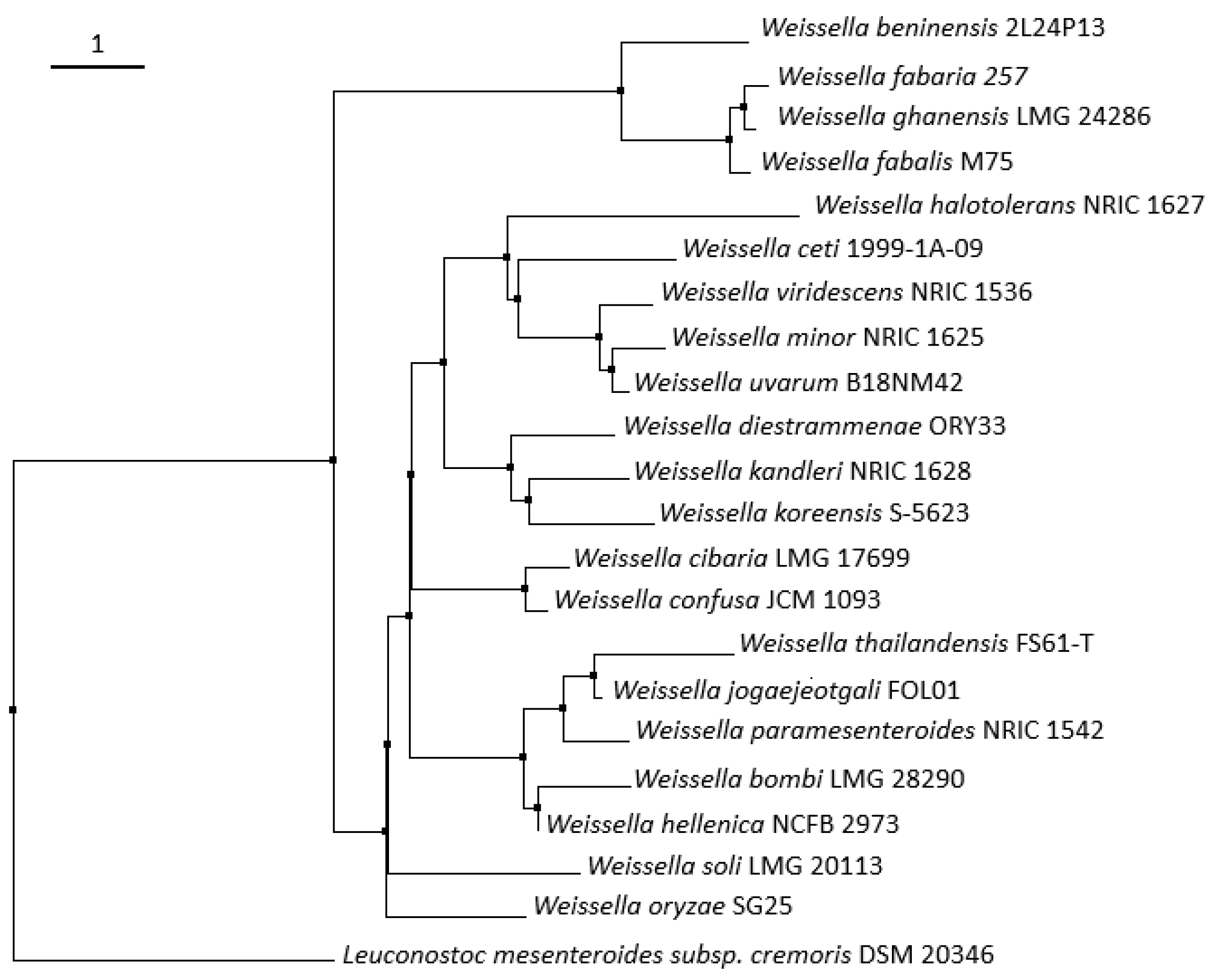
| Weissella spp. | Sources | Genome Size (Mb) | Taxonomic Branch | References |
|---|---|---|---|---|
| W. fabalis | Cocoa bean fermentation | - | 1 | [26] |
| W. fabaria | Cocoa bean fermentation | - | 1 | [27] |
| W. ghanensis | Cocoa bean fermentation | - | 1 | [28] |
| W. beninensis | Fermented cassava | - | 1 | [16] |
| W. jogaejeotgali | Jeotgal (fermented seafood) | 2.11 (CP014332.1) | 2 | [29] |
| W. thailandensis | Jeotgal, Mexican cheese, Thai fermented meat and fish | 1.97 (HE575133 to HE575182) | 2 | [30]; See Table 2 |
| W. paramesenteroides | Fermented sausages, fermented fruits and vegetables, fermented milk, cheese, fermented cassava, sourdough, fermented cereal, fermented fish | 1.96 (NZ_ACKU00000000.1) | 2 | [31,32]; See Table 2 |
| W. hellenica | Fermented sausages, fermented vegetables, fermented milk, cheese, fermented maize beverage, fermented meat | 1.82–1.92 (NZ_BBIK00000000.1; NZ_FMAW00000000.1) | 2 | [19]; See Table 2 |
| W. bombi | Gut of a bumble bee | - | 2 | [33] |
| W. halotolerans | Meat products, fermented sea food, cheese, fermented sausages | 1.36 (NZ_ATUU00000000.1) | 3 | [20]; See Table 2 |
| W. ceti | Beaked whale | 1.35–1.39 (NZ_ANCA00000000.1; NZ_CP009224.1) | 3 | [17] |
| W. viridescens | Meat products, dry salami, kimchi, fermented milk, cheese, fermented cereal beverage, fermented sausages, fermented fish | 1.53–1.56 (NZ_JQBM00000000.1; NZ_CYXF00000000.1) | 3 | [24]; See Table 2 |
| W. minor | Sludge of milking machine, fermented fruits and vegetables, fermented dry salami | 1.77 (NZ_JQCD00000000.1) | 3 | [20]; See Table 2 |
| W. uvarum | Wine grapes | - | 3 | [25] |
| W. diestrammenae | Gut of a camel cricket | - | 4 | [34] |
| W. kandleri | Desert spring and desert plants | 1.33 (NZ_JQBP00000000.1) | 4 | [35] |
| W. koreensis | Kimchi, fermented vegetables | 1.42–1.73 (NC_015759.1; NZ_AKGG00000000.1) | 4 | [36]; See Table 2 |
| W. cibaria | Malaysian ingredients foods, clinical samples, animals, human feces, fermented fruits and vegetables, fermented dairy products, cheese, fermented cassava, fermented cereal foods and beverages, sourdough, cocoa bean fermentation, fermented sausages, fermented fish | 2.32–2.47 (NZ_AEKT00000000.1; NZ_CP012873.1) | 5 | [37]; See Table 2 |
| W. confusa | Sugar cane, animal, clinical samples, Malaysian ingredients foods, human feces, pasta manufacturing, kimchi, fermented vegetables, fermented milk, cheese, fermented cereal foods, fermented cassava, sourdough, cocoa bean fermentation, pork sausages, fermented fish | 2.18–2.28 (NZ_MNBZ00000000.1; NZ_CAGH00000000.1) | 5 | [37,38]; See Table 2 |
| W. oryzae | Fermented rice grains, cereal based beverage | 2.13 (NZ_BAWR00000000.1) | 0 | [34,39] |
| W. soli | Soil, kimchi, Jeotgal, stinky tofu, leek fermentation, sliced cabbage | - | 0 | [40]; See Table 2; [31] |
| Food Category | Fermented Product | Weissella Species | Reference |
|---|---|---|---|
| Fruits and Vegetables Fermented Food | Kimchi (mix vegetables) | W. koreensis, W. cibaria, W. confusa, W. soli, W. hellenica | [36,41,42,43,44,45,46] |
| Yan-dong-gua (wax gourd) | W. cibaria, W. paramesenteroides | [47] | |
| Pobuzihi (cummingcordia) | W. cibaria, W. paramesenteroides | [48] | |
| Jiang-gua (cucumbers) | W. cibaria, W. hellenica | [49] | |
| Yan-tsai-shin (brocoli stems) | W. cibaria, W. paramesenteroides, W. minor | [50] | |
| Yan-Jiang (ginger) | W. cibaria | [51] | |
| Yan-taozih (pickled peaches) | W. cibaria, W. paramesenteroides, W. minor | [52] | |
| Xi-gua-mian (watermelon) | W. paramesenteroides | [53] | |
| Dochi (black beans) | W. paramesenteroides | [54] | |
| Koozh (cucumber) | W. koreensis | [55] | |
| Suan-tsai, fu-tsai (mustard product) | W. cibaria, W. paramesenteroides | [56] | |
| Jeotgal (sea food) | W. thailandensis, W. halotolerans, W. soli, W. cibaria, W. jogaejeotgali | [29,43] | |
| Stinky tofu | W. cibaria, W. confusa, W. paramesenteroides, W. soli | [57] | |
| Tuaw jaew (soybeans) | W. confusa | [58] | |
| Fermented sea food | W. paramesenteroides | [59] | |
| Fermented cabbage | W. cibaria | [60] | |
| Tempoyak (durian) | W. paramesenteroides | [61] | |
| Commercial cucumber fermentation | W. cibaria, W. paramesenteroides | [62] | |
| Cauliflower and mixed vegetables | W. cibaria, W. paramesenteroides | [63] | |
| Well-fermented leek kimchi | W. viridescens, W. confusa, W. cibaria | [64] | |
| Leek fermentation | W. soli, Weissella spp. | [65] | |
| Commercial sauerkraut | Weissella spp. | [66] | |
| Sauerkraut | Weissella confusa/cibaria | [67] | |
| Dairy Fermented Food | Nono (Fermented skimmed milk), cheese | W. confusa | [68] |
| Nunu (Ghanaian fermented milk) | W. confusa | [69] | |
| Kule naoto (Masain fermented milk in Kenya) | W. paramesenteroides/Lc mesenteroides | [70] | |
| Maisian fermented zebu milk | W. confusa (Lb confusus), W. viridescens (Lb viridescens) | [71] | |
| Mongolian fermented milk | W. viridescens | [72] | |
| Shubat (fermented camel milk) | W. hellenica | [73] | |
| Dahi (Indian yogurt like product) | W. cibaria | [60] | |
| Italian cheese | W. confusa, W. cibaria | [74] | |
| Mexican cheese (Cotija cheese) | W. thailandensis | [75] | |
| Manura (hard cheese) | W. paramesenteroides, W. viridescens | [76] | |
| Scamorza Altamurana Cheese | W. viridescens | [77] | |
| Mozarella cheese | W. hellenica | [78] | |
| Cheddar cheese | W. halotolerans, W. viridescens | [79] | |
| Romanian cheese | W. viridescens | [80] | |
| Mongolian dairy products | W. cibaria | [81] | |
| Starchy or Cereal-Based Fermented Food and Diverse | Togwa (sorghum based food) | W. confusa | [82] |
| Gari, Attieke, Lafun (cassava) | W. paramesenteroides, W. cibaria, W. confusa, W. beninensis | [16,83,84,85,86] | |
| Idli batter (Indian fermented rice and black gram based food) | W. confusa, W. cibaria | [60,87] | |
| French wheat sourdough, buckwheat and teff sourdough, Italian sourdough, Turkish sourdough, spontaneous sourdough | W. confusa, W. cibaria, W. paramesenteroides | [10,88,89,90,91,92,93] | |
| Mexican Pozol | W. paramesenteroides | [94] | |
| Fermented rice grains | W. oryzae | [95] | |
| Chicha (maize based beverage) | W. confusa, W. hellenica, W. paramesenteroides | [96] | |
| Kunu-zaki (Nigerian cereal based food) | W. confusa | [97] | |
| Boza (Bulgarian cereal-based beverage) | W. confusa, W. oryzae | [39] | |
| Fura (African millet based food) | W. confusa | [98] | |
| Borde (Ethiopian cereal beverage) | W. confusa, W. viridescens | [99] | |
| Cocoa bean fermentation | W. cibaria, W. ghanensis, W. confusa, W. paramesenteroides, W. fabaria, W. fabalis | [26,27,28,100] | |
| Makgeolli (Korean starchy alcoholic beverage) | W. confusa, W. cibaria, W. paramesenteroides | [101] | |
| Fermented Meat and Fish Product | Nham (Thai fermented pork sausage) | W. cibaria, W. confusa | [102,103,104] |
| Italian fermented sausages | W. hellinica, W. paramesenteroides | [105,106] | |
| Greek dry-fermented sausages | W. viridescens | [107] | |
| Fermented sausages in Hungary | W. paramesenteroides/W. hellenica, W. viridescens | [108] | |
| Alheira (Fermented sausage in Portugal) | W. cibaria, W. viridescens | [109] | |
| Sucuk (fermented turkish sausages) | W. viridescens | [110] | |
| Portuguese fermented sausage | W. halotolerans | [111] | |
| Smoked horsemeat sausage | W. hellenica, Weissella spp. | [112] | |
| Fermented sausages | W. hellenica | [19] | |
| Fermented Greek dry salami | W. hellenica, W. viridescens, W. paramesenteroides, W. minor, W. halotolerans | [113] | |
| Chinese dong fermented meat | W. hellenica | [114] | |
| Mum (Thai fermented meat) | W. thailandensis | [58] | |
| Pla-ra, Pla-ra sub, plaa-som, pla-jom, pla-jaw (Thai fermented fish) | W. thailandensis, W. cibaria, W. confusa, W. paramesenteroides, W. viridescens | [30,58,115,116,117] | |
| Sidra (Fish products) | W. confusa | [118] |
| Type of Fermented Product | Product | LAB Species Used as Commercial Starters | References |
|---|---|---|---|
| Dairy Fermented Foods | Yoghurt | S. thermophilus, Lb. delbruecki subsp. bulgaricus | [130] |
| Fermented/Probiotic milk | Lb. casei, Lb. acidophilus, Lb. rhamnosus, Lb. johnsonii, B. lactis, B. bifidum, B. brevis | ||
| Kefir | Lb. kefir, Lb. kefiranofacies, Lb. brevis | ||
| Butter and buttermilk | L. lactis susbp. lactis, L. lactis subsp. diacetylactis, L. lactis subsp. cremoris, Lc. mesenteroides subsp. cremoris | ||
| Swiss and Italian type cheeses | Lb. delbrueckii subsp. lactis, Lb. helveticus, Lb. casei, Lb. delbrueckii subsp. bulgaricus, S. thermophilus | ||
| Cheeses (with or without eyes) | L. lactis subsp. Lactis, L. lactis subsp. cremoris, L. lactis subsp. diacetylactis, Lc. mesenteroides subsp. cremoris | ||
| Fermented Cereals | Sourdough | Lb. brevis, Lb. plantarum, Lb. sanfranciscensis, Lb. casei, Lb. delbrueckii, Lb. fermentum, P. pentosaceus, P. acidilactic, Lb. pontis, Lb. crispatus, Lb. paracasei, Lb. helveticus, Lb. paralimentarius, Lc. lastis | [6] |
| Fruits and Vegetables | Sauerkraut | Lc. mesenteroides, Lb. plantarum, P. acidilactici | [8] |
| Pickles | Lc. mesenteroides, P. cerevisiae, Lb. brevis, Lb. plantarum | ||
| Fermented olives | Lb. paracasei, Lb. pentosus, Lb. plantarum | ||
| Fermented vegetables | P. acidilactici, P. pentosaceus, Lb. plantarum, Lb. fermentum | ||
| Vegetable juices | Lb. acidophilus, Lb. bavaricus, Lb. bifidus, Lb. brevis, Lb. casei, Lb. delbrueckii, Lb. helveticus, Lb. plantarum, Lb. salivarius, Lb. xylosus, L. lactis, Lc. mesenteroides | [3] | |
| Meat Products | Sausages | P. acidilactici, P. pentosaceus, Lb. sakei, Lb. curvatus, Lb. plantarum, Lb. pentosus, Lb, casei, L. lactis | [124,131] |
| Fermented Beverages | Wine | Oenococcus oeni, Lb. plantarum, Lb. hilgardii | [3] |
| Criteria Category | Fruits and Vegetables | Dairy Products | Meat Products | Cereal Based Foods |
|---|---|---|---|---|
| Technological | - Growth and acidification rate - Salt tolerance - Tolerance to low values of pH - Growth at low temperature - Completeness of fermentation - Malolatic fermentation - Tolerance to phenols - Synthesis of antimicrobial substances - No formation of hydrogen peroxide - Pectinolytic activity | - Growth and acidification rate - Production of nutraceuticals - Accelerate ripening of cheese - Resistance to bacteriophage - Proteinase and peptidase activity | - Fast production of lactic acid - Growth rate at different temperatures - Salt and pH tolerance - Persistence over the whole fermentation and ripening process - Nitrate and nitrite reduction - Catalase positive - Lactose negative - Proteolytic and lipolytic enzyme activities - No formation of hydrogen peroxide - Antagonism against pathogens - Improve the nutritional value of the sausages | - Growth and acidification rate - Salt tolerance - Growth at low temperatures - Synthesis of antimicrobial compounds |
| Sensory | - Hetero-fermentative metabolism - Synthesis of aroma compounds and their precursors | - Production of aroma and flavor - Synthesis of exopolysaccharides | - Formation of flavor - Hetero-fermentative metabolism | - Hetero-fermentative metabolism - Synthesis of aroma compounds and their precursors |
| Nutritional | - Synthesis of exopolysaccharides - Increase of the antioxidant activity - Synthesis of biogenic compounds - Bacteriocin production | - Synthesis of exo-polysaccharides - Bacteriocins production - Reduction of toxic or antinutritional factors - Low-calorie sugar production - Vitamin production - Bioactive peptide production - Production of conjugated linolein acid (CLA) | - Tolerance or even synergy to other microbial components or starters - No formation of ropy slime - Bacteriocin production - Probiotic features | - Release of free amino acids - Synthesis of biogenic compounds - Degradation of antinutritionnal factor (phytic acid) - Increase the antioxidant activity - Synthesis of exopolysaccharides |
| Safety | - No formation of biogenic amines - No antibiotic resistance profile | |||
| Species | Source | Type of EPS | Molecular Mass (Da) | Amount Produced | Media | Reference |
|---|---|---|---|---|---|---|
| W. cibaria (WC4) | Sourdough | Glucan | 1.1–1.3 × 104 | 7.9 g·L−1 (6 d) | MRS + Sucrose | [10] |
| 2.5 g·kg−1 | Sourdough | |||||
| W. cibaria (WC3) | Sourdough | Glucan | 1.1–1.3 × 104 | 6.7 g·L−1 (6 d) | MRS + Sucrose | [10] |
| W. cibaria (WC9) | Sourdough | Glucan | 1.1–1.3 × 104 | 5.5 g·L−1 (6 d) | MRS + Sucrose | [10] |
| W. cibaria (MG1) | Sourdough | Dextran | 106 to 107 | 0.9 g·kg−1 | Sourdough | [149] |
| 3.2 g·kg−1 | ||||||
| 4.2 g·kg−1 | ||||||
| 7.2 × 108 | 8 g·kg−1 | [151] | ||||
| 36.4 g·L−1 | MRS + Sucrose | [153] | ||||
| W. cibaria (11GM-2) | Sour milk | Dextran | >2 × 107 | ND | MRS + Sucrose | [152] |
| W. cibaria (CMGDEX3) | Cabbage | Dextran | >2 × 106 | 0.24 g·L−1 | MRS + Sucrose | [154] |
| W. cibaria (JAG8) | Apple | Dexran | 8 × 105 | 38 g·L−1 (12 h) | Tsuchiya medium | [155,156] |
| W. confusa (E392) | Soured carrot mash | Dextran | ND | ND | MRS + Sucrose | [157] |
| W. confusa (KR780676) | Idli batter | Galactan | ND | 17.2 g·kg−1 (dry weight) | MRS + Sucrose | [87] |
| W. confusa (F3/2-2) | Cassava fermentation | Dextran | >2 × 107 | ND | MRS + Sucrose | [152] |
| Levan | 2 × 105 | ND | MRS + Raffinose | |||
| W. confusa (8CS-2) | Sour milk | HePS | ND | ND | MRS + Glucose | [152] |
| Dextran | >2 × 107 | ND | MRS + Sucrose | |||
| W. confusa (NH 02) | Nham | ND | 1.13 × 106 | 18.08 g·L−1 | MRS + Sucrose | [104] |
| W. hellenica (SKkimchi3) | Kimchi | Glucan | 2.03 × 105 | 5.12 g·L−1 | MRS + Sucrose | [45] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fessard, A.; Remize, F. Why Are Weissella spp. Not Used as Commercial Starter Cultures for Food Fermentation? Fermentation 2017, 3, 38. https://doi.org/10.3390/fermentation3030038
Fessard A, Remize F. Why Are Weissella spp. Not Used as Commercial Starter Cultures for Food Fermentation? Fermentation. 2017; 3(3):38. https://doi.org/10.3390/fermentation3030038
Chicago/Turabian StyleFessard, Amandine, and Fabienne Remize. 2017. "Why Are Weissella spp. Not Used as Commercial Starter Cultures for Food Fermentation?" Fermentation 3, no. 3: 38. https://doi.org/10.3390/fermentation3030038





