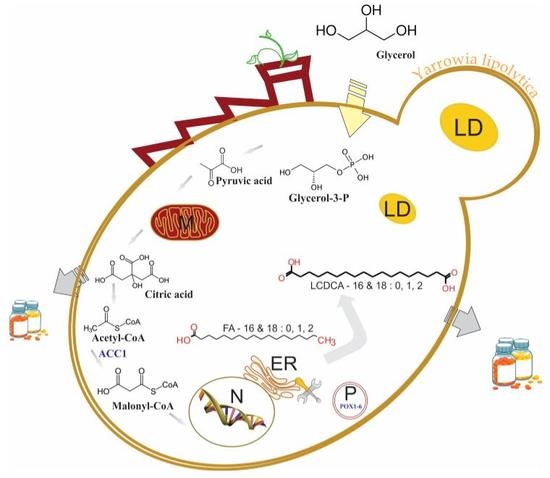
Combinatorial Engineering of Yarrowia lipolytica as a Promising Cell Biorefinery Platform for the de novo Production of Multi-Purpose Long Chain Dicarboxylic Acids
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Batch Fermentation
2.3. Genetic Techniques
2.4. Analytical Methods
2.4.1. Dry Biomass
2.4.2. Glycerol and Citric Acid Concentrations
2.4.3. Qualitative and Quantitative Analysis of Fatty Acids and LCDCAs
3. Results
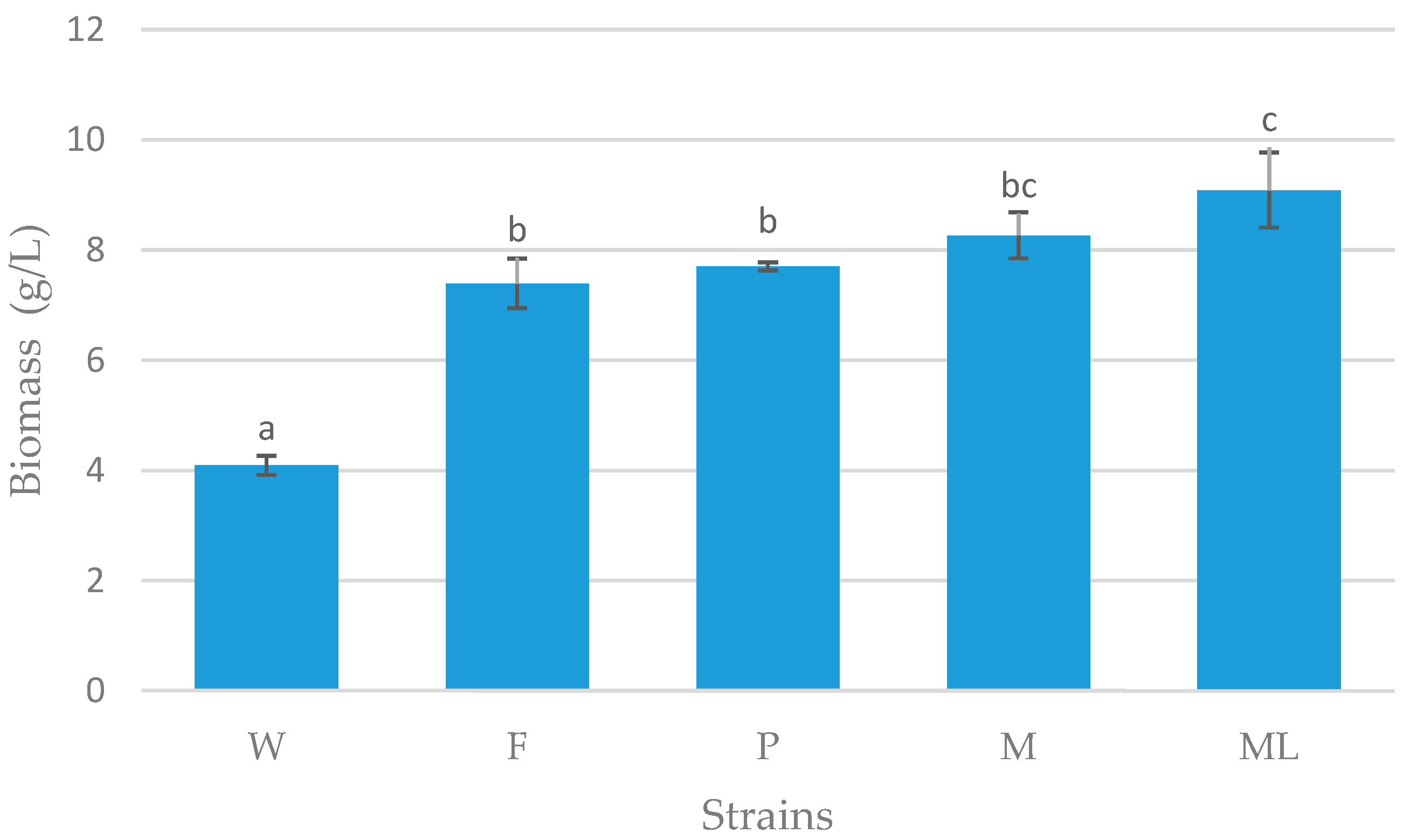
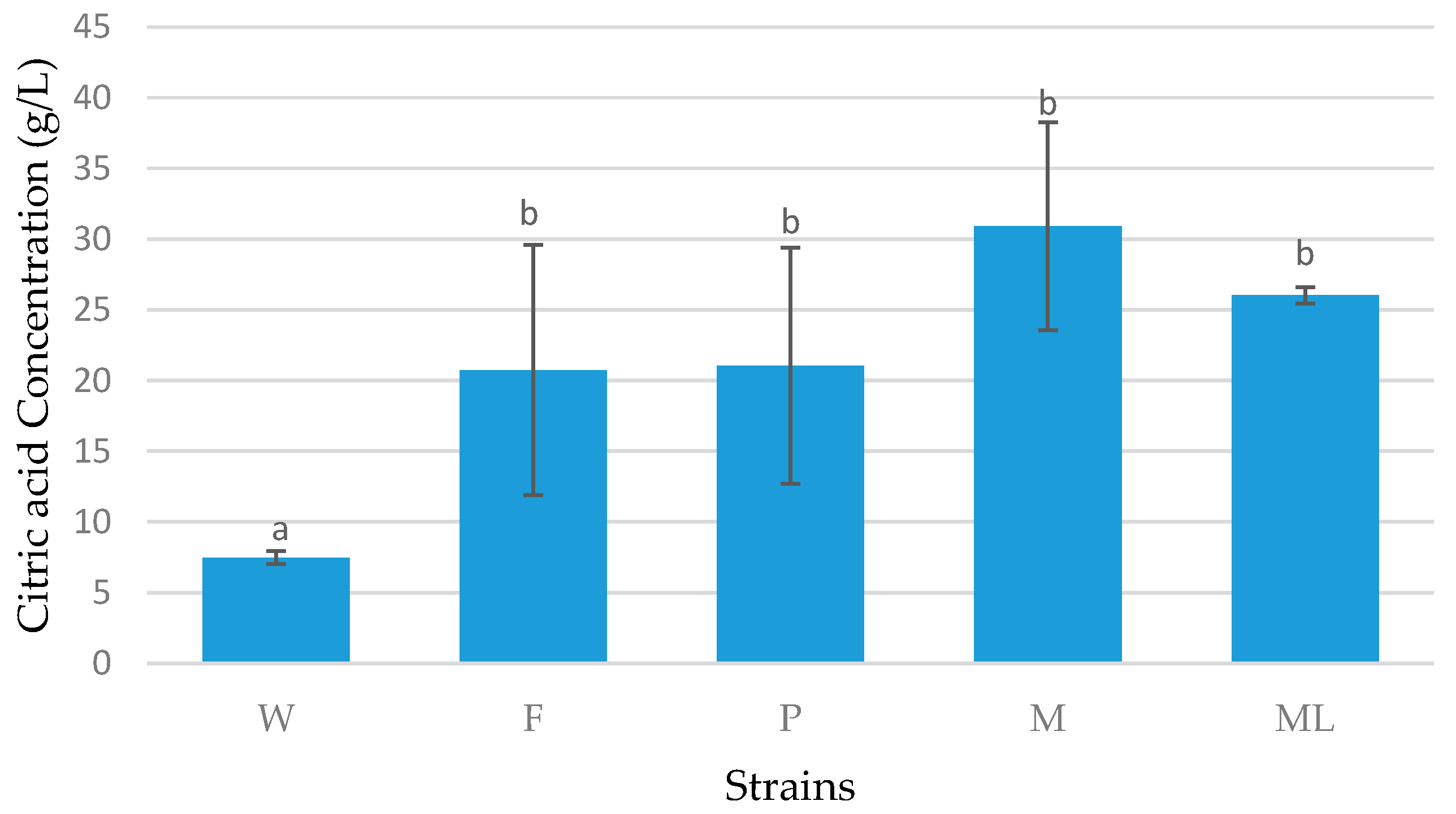
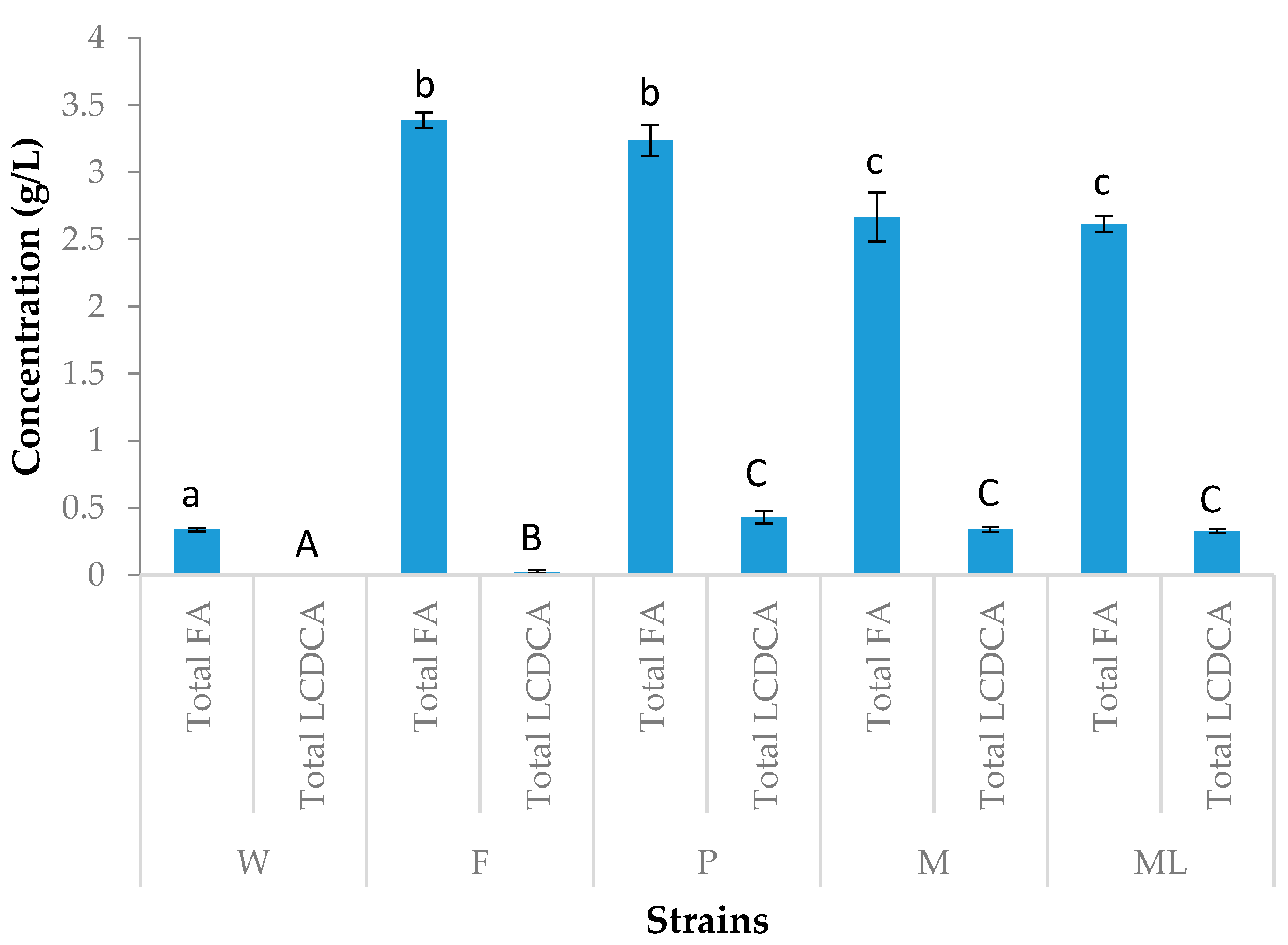
3.1. Screening Process
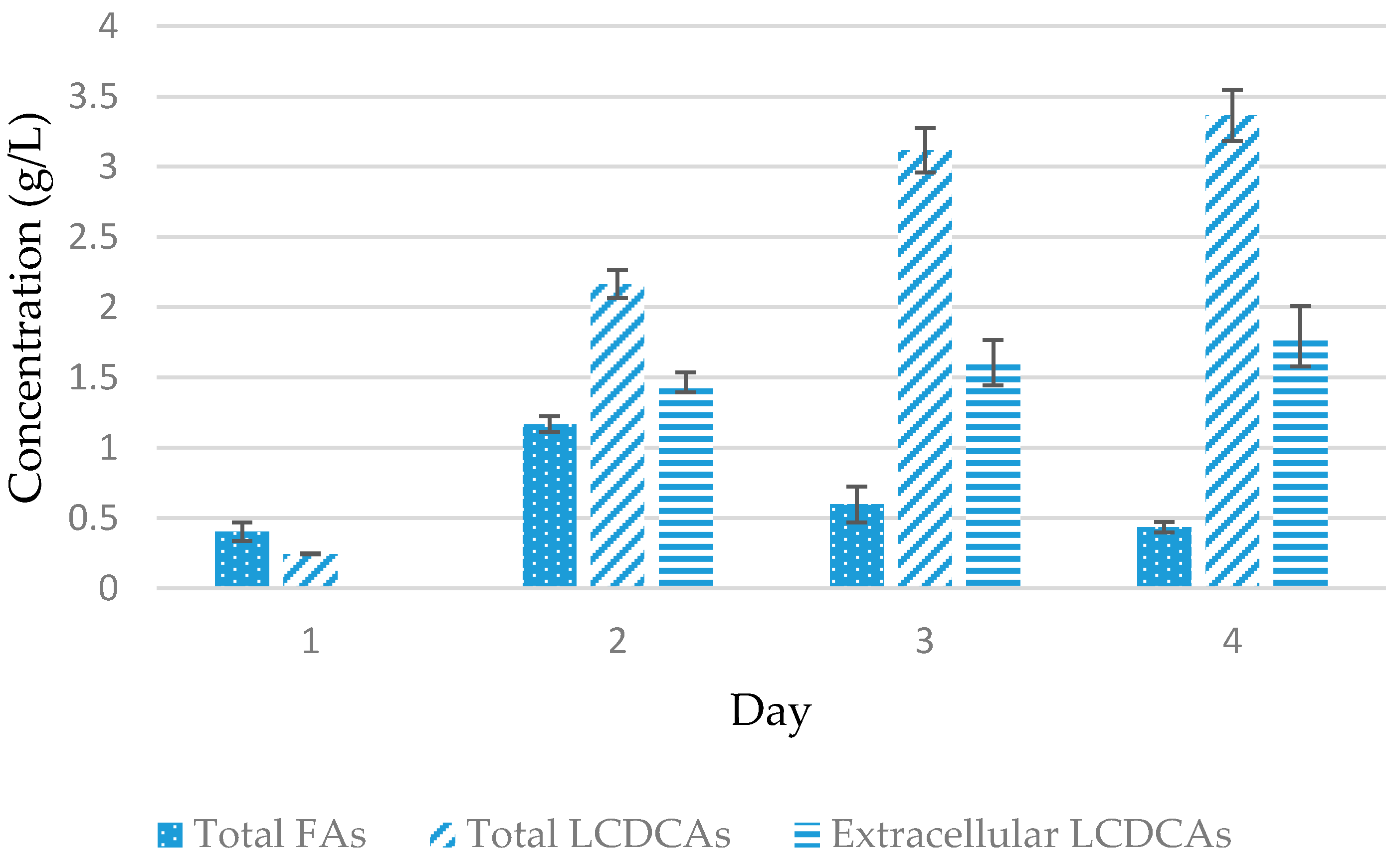
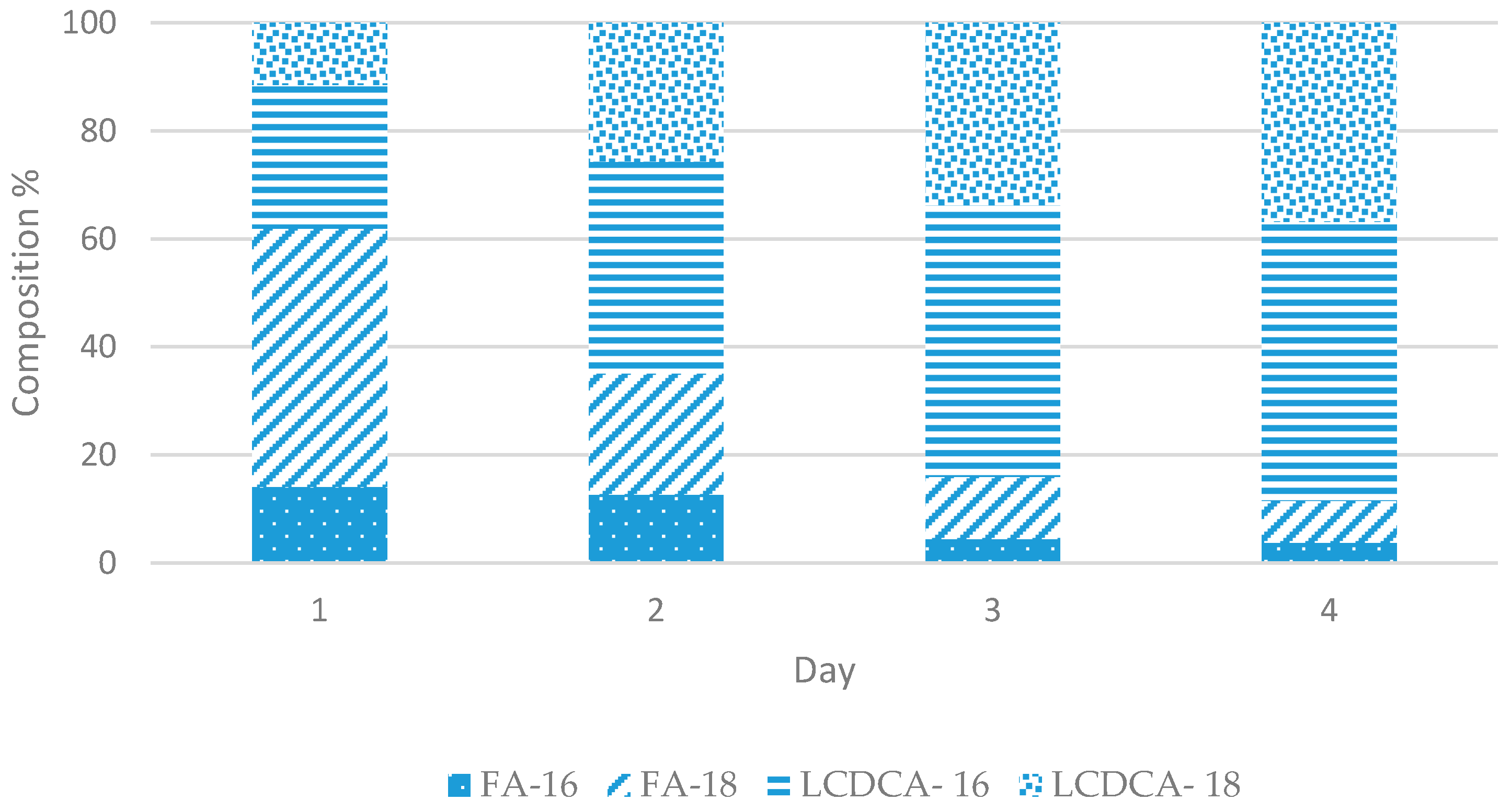
3.2. Preliminary Shake Flask Experiments for Fatty Acid and LCDCA Production
3.3. Batch Fermentation
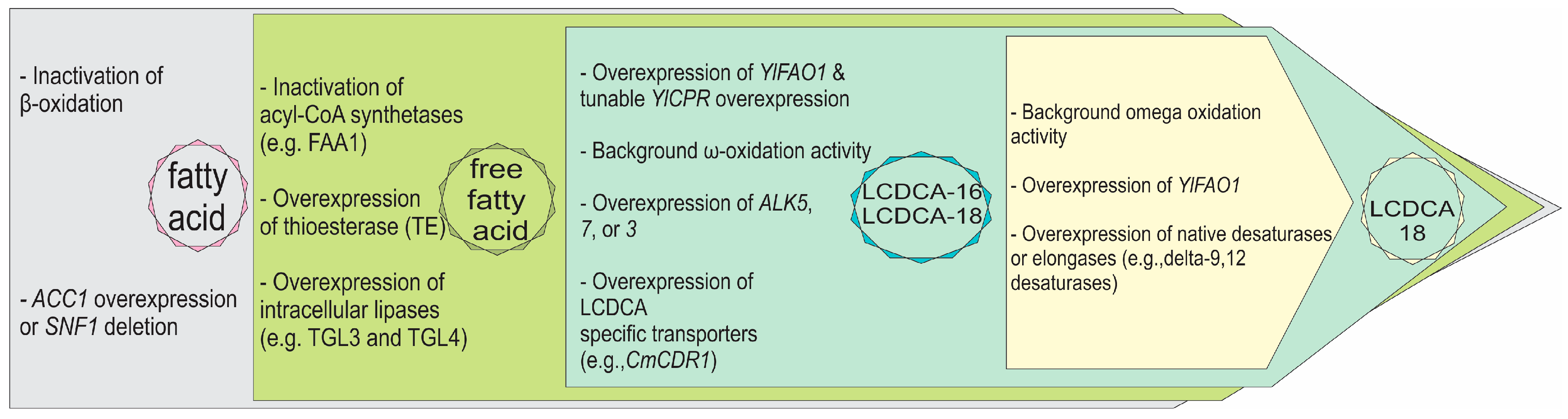
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pellis, A.; Herrero Acero, E.; Gardossi, L.; Ferrario, V.; Guebitz, G.M. Renewable building blocks for sustainable polyesters: New biotechnological routes for greener plastics. Polym. Int. 2016, 65, 861–871. [Google Scholar] [CrossRef]
- Liao, J.C.; Mi, L.; Pontrelli, S.; Luo, S. Fuelling the future: Microbial engineering for the production of sustainable biofuels. Nat. Rev. Microbiol. 2016, 14, 288–304. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.; Yang, J.E.; Ha, J.Y.; Chae, T.U.; Shin, J.H.; Gustavsson, M.; Lee, S.Y. Bio-based production of monomers and polymers by metabolically engineered microorganisms. Curr. Opin. Biotechnol. 2015, 36, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Díaz, A.; Katsarava, R.; Puiggalí, J. Synthesis, properties and applications of biodegradable polymers derived from diols and dicarboxylic acids: From polyesters to poly (ester amide) s. Int. J. Mol. Sci. 2014, 15, 7064–7123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beuhler, A. C18 Diacid Market to Grow and Expand into an Array of Novel Products with Superior Properties. 2013. Available online: http://www.elevance.com/images/documents/Elevance-ODDA-C18-white-paper_20130916_F.pdf (accessed on 22 July 2017).
- Bowen, C.H.; Bonin, J.; Kogler, A.; Barba-Ostria, C.; Zhang, F. Engineering Escherichia coli for conversion of glucose to medium-chain ω-hydroxy fatty acids and α, ω-dicarboxylic acids. ACS Synth. Biol. 2015, 5, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Sathesh-Prabu, C.; Lee, S.K. Production of long-chain α, ω-dicarboxylic acids by engineered escherichia coli from renewable fatty acids and plant oils. J. Agric. Food Chem. 2015, 63, 8199–8208. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Ness, J.E.; Xie, W.; Zhang, X.; Minshull, J.; Gross, R.A. Biosynthesis of monomers for plastics from renewable oils. J. Am. Chem. Soc. 2010, 132, 15451–15455. [Google Scholar] [CrossRef] [PubMed]
- Ngo, H.L.; Foglia, T.A. Synthesis of long chain unsaturated-α, ω-dicarboxylic acids from renewable materials via olefin metathesis. J. Am. Oil Chem. Soc. 2007, 84, 777–784. [Google Scholar] [CrossRef]
- Huf, S.; Kruegener, S.; Hirth, T.; Rupp, S.; Zibek, S. Biotechnological synthesis of long-chain dicarboxylic acids as building blocks for polymers. Eur. J. Lipid Sci. Technol. 2011, 113, 548–561. [Google Scholar] [CrossRef]
- Cho, C.; Choi, S.Y.; Luo, Z.W.; Lee, S.Y. Recent advances in microbial production of fuels and chemicals using tools and strategies of systems metabolic engineering. Biotechnol. Adv. 2015, 33, 1455–1466. [Google Scholar] [CrossRef] [PubMed]
- Weiss, A. Selective microbial oxidations in industry: Oxidations of alkanes, fatty acids, heterocyclic compounds, aromatic compounds and glycerol using native or recombinant microorganisms. Mod. Biooxid. Enzym. React. Appl. 2007, 193–209. [Google Scholar] [CrossRef]
- Kroha, K. Industrial biotechnology provides opportunities for commercial production of new long-chain dibasic acids. Inform 2004, 15, 568–571. [Google Scholar]
- Picataggio, S.; Rohrer, T.; Deanda, K.; Lanning, D.; Reynolds, R.; Mielenz, J.; Eirich, L.D. Metabolic engineering of candida tropicalis for the production of long–chain dicarboxylic acids. Nat. Biotechnol. 1992, 10, 894–898. [Google Scholar] [CrossRef]
- Maurer, S.C.; Schmid, R.D. Biocatalysts for the epoxidation and hydroxylation of fatty acids and fatty alcohols. Handb. Ind. Biocatal. 2005. [Google Scholar] [CrossRef]
- Kogure, T.; Horiuchi, H.; Matsuda, H.; Arie, M.; Takagi, M.; Ohta, A. Enhanced induction of cytochromes p450alk that oxidize methyl-ends of n-alkanes and fatty acids in the long-chain dicarboxylic acid-hyperproducing mutant of candida maltosa. FEMS Microbiol. Lett. 2007, 271, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Cryle, M.J.; De Voss, J.J.; de Montellano, P.R.O. Functional expression and characterization of cytochrome p450 52a21 from candida albicans. Arch. Biochem. Biophys. 2007, 464, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Scheps, D.; Honda Malca, S.; Richter, S.M.; Marisch, K.; Nestl, B.M.; Hauer, B. Synthesis of ω-hydroxy dodecanoic acid based on an engineered cyp153a fusion construct. Microb. Biotechnol. 2013, 6, 694–707. [Google Scholar] [CrossRef] [PubMed]
- Girhard, M.; Bakkes, P.J.; Mahmoud, O.; Urlacher, V.B. P450 biotechnology. In Cytochrome p450; Springer: Berlin, Gemany, 2015; pp. 451–520. [Google Scholar]
- Li, H.; Pinot, F.; Sauveplane, V.; Werck-Reichhart, D.; Diehl, P.; Schreiber, L.; Franke, R.; Zhang, P.; Chen, L.; Gao, Y. Cytochrome p450 family member cyp704b2 catalyzes the ω-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 2010, 22, 173–190. [Google Scholar] [CrossRef] [PubMed]
- Malca, S.H.; Scheps, D.; Kühnel, L.; Venegas-Venegas, E.; Seifert, A.; Nestl, B.M.; Hauer, B. Bacterial cyp153a monooxygenases for the synthesis of omega-hydroxylated fatty acids. Chem. Commun. 2012, 48, 5115–5117. [Google Scholar] [CrossRef] [PubMed]
- Höfer, R.; Briesen, I.; Beck, M.; Pinot, F.; Schreiber, L.; Franke, R. The arabidopsis cytochrome p450 cyp86a1 encodes a fatty acid ω-hydroxylase involved in suberin monomer biosynthesis. J. Exp. Bot. 2008, 59, 2347–2360. [Google Scholar] [CrossRef] [PubMed]
- Kandel, S.; Sauveplane, V.; Compagnon, V.; Franke, R.; Millet, Y.; Schreiber, L.; Werck-Reichhart, D.; Pinot, F. Characterization of a methyl jasmonate and wounding-responsive cytochrome p450 of arabidopsis thaliana catalyzing dicarboxylic fatty acid formation in vitro. FEBS J. 2007, 274, 5116–5127. [Google Scholar] [CrossRef] [PubMed]
- Benveniste, I.; Tijet, N.; Adas, F.; Philipps, G.; Salaün, J.-P.; Durst, F. Cyp86a1 fromarabidopsis thalianaencodes a cytochrome p450-dependent fatty acid omega-hydroxylase. Biochem. Biophys. Res. Commun. 1998, 243, 688–693. [Google Scholar] [CrossRef] [PubMed]
- Craft, D.L.; Madduri, K.M.; Eshoo, M.; Wilson, C.R. Identification and characterization of the cyp52 family of candida tropicalis atcc 20336, important for the conversion of fatty acids and alkanes to alpha,omega-dicarboxylic acids. Appl. Environ. Microbiol. 2003, 69, 5983–5991. [Google Scholar] [CrossRef] [PubMed]
- Eschenfeldt, W.H.; Zhang, Y.; Samaha, H.; Stols, L.; Eirich, L.D.; Wilson, C.R.; Donnelly, M.I. Transformation of fatty acids catalyzed by cytochrome p450 monooxygenase enzymes of candida tropicalis. Appl. Environ. Microbiol. 2003, 69, 5992–5999. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, S.M.; Danesh-Azari, H.R.; Spandolf, C.; Weissenborn, M.J.; Grogan, G.; Hauer, B. Structure-guided redesign of CYP153AM. aq for the improved terminal hydroxylation of fatty acids. ChemCatChem 2016, 8, 3234–3239. [Google Scholar] [CrossRef]
- Schreiber, L.; Durst, F.; Pinot, F. Cyp94a5, a new cytochrome p450 from nicotiana tabacum is able to catalyze the oxidation of fatty acids to the v-alcohol and to the corresponding diacid. FEBS J. 2001, 268, 3083–3090. [Google Scholar]
- McLean, K.J.; Luciakova, D.; Belcher, J.; Tee, K.L.; Munro, A.W. Biological diversity of cytochrome p450 redox partner systems. In Monooxygenase, Peroxidase and Peroxygenase Properties and Mechanisms of Cytochrome p450; Springer: Berlin, Gemany, 2015; pp. 299–317. [Google Scholar]
- Girhard, M.; Tieves, F.; Weber, E.; Smit, M.S.; Urlacher, V.B. Cytochrome p450 reductase from candida apicola: Versatile redox partner for bacterial p450s. Appl. Microbiol. Biotechnol. 2013, 97, 1625–1635. [Google Scholar] [CrossRef] [PubMed]
- Theron, C.W. Hetrologous Expression of Cytochrome p450 Monoxygenase by the Yeast Yarrowia Lipolytica; University of the Free State: Bloemfontein, South Africa, 2007. [Google Scholar]
- Iida, T.; Sumita, T.; Ohta, A.; Takagi, M. The cytochrome p450alk multigene family of an n-alkane-assimilating yeast, yarrowia lipolytica: Cloning and characterization of genes coding for new CYP52 family members. Yeast 2000, 16, 1077–1087. [Google Scholar] [CrossRef]
- Abghari, A.; Chen, S. Yarrowia lipolytica as an oleaginous cell factory platform for the production of fatty acid-based biofuel and bioproducts. Front. Energy Res. 2014, 2. [Google Scholar] [CrossRef]
- Juretzek, T.; Mauersberger, S.; Barth, G. Recombinant Haploid or Diploid Yarrowia Lipolytica Cells for the Functional Heterologous Expression of Cytochrome p450 Systems. Patent WO2000003008 A3, 27 April 2000. [Google Scholar]
- Fickers, P.; Benetti, P.H.; Waché, Y.; Marty, A.; Mauersberger, S.; Smit, M.S.; Nicaud, J.M. Hydrophobic substrate utilisation by the yeast yarrowia lipolytica, and its potential applications. FEMS Yeast Res. 2005, 5, 527–543. [Google Scholar] [CrossRef] [PubMed]
- Iwama, R.; Kobayashi, S.; Ishimaru, C.; Ohta, A.; Horiuchi, H.; Fukuda, R. Functional roles and substrate specificities of twelve cytochromes p450 belonging to cyp52 family in n-alkane assimilating yeast yarrowia lipolytica. Fungal Genet. Biol. 2016, 91, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Braun, A.; Geier, M.; Buehler, B.; Schmid, A.; Mauersberger, S.; Glieder, A. Steroid biotransformations in biphasic systems with yarrowia lipolytica expressing human liver cytochrome p450 genes. Microb. Cell Factories 2012, 11, 106. [Google Scholar] [CrossRef] [PubMed]
- Mauersberger, S. Cytochromes p450 of the alkane-utilising yeast yarrowia lipolytica. In Yarrowia Lipolytica; Springer: Berlin, Gemany, 2013; pp. 227–262. [Google Scholar]
- Takai, H.; Iwama, R.; Kobayashi, S.; Horiuchi, H.; Fukuda, R.; Ohta, A. Construction and characterization of a yarrowia lipolytica mutant lacking genes encoding cytochromes p450 subfamily 52. Fungal Genet. Biol. 2012, 49, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Thevenieau, F.; Beopoulos, A.; Desfougeres, T.; Sabirova, J.; Albertin, K.; Zinjarde, S.S.; Nicaud, J.-M. Uptake and assimilation of hydrophobic substrates by the oleaginous yeast yarrowia lipolytica. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin/Heidelberg, Gemany, 2010; pp. 1513–1527. [Google Scholar]
- Hanley, K.; Nguyen, L.V.; Khan, F.; Pogue, G.P.; Vojdani, F.; Panda, S.; Pinot, F.; Oriedo, V.B.; Rasochova, L.; Subramanian, M. Development of a plant viral-vector-based gene expression assay for the screening of yeast cytochrome p450 monooxygenases. Assay Drug Dev. Technol. 2003, 1, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Gatter, M.; Förster, A.; Bär, K.; Winter, M.; Otto, C.; Petzsch, P.; Ježková, M.; Bahr, K.; Pfeiffer, M.; Matthäus, F. A newly identified fatty alcohol oxidase gene is mainly responsible for the oxidation of long-chain ω-hydroxy fatty acids in yarrowia lipolytica. FEMS Yeast Res. 2014, 14, 858–872. [Google Scholar] [CrossRef] [PubMed]
- Iwama, R.; Kobayashi, S.; Ohta, A.; Horiuchi, H.; Fukuda, R. Alcohol dehydrogenases and an alcohol oxidase involved in the assimilation of exogenous fatty alcohols in yarrowia lipolytica. FEMS Yeast Res. 2015, 15, fov014. [Google Scholar] [CrossRef] [PubMed]
- Beopoulos, A.; Chardot, T.; Nicaud, J.-M. Yarrowia lipolytica: A model and a tool to understand the mechanisms implicated in lipid accumulation. Biochimie 2009, 91, 692–696. [Google Scholar] [CrossRef] [PubMed]
- Smit, M.S.; Mokgoro, M.M.; Setati, E.; Nicaud, J.-M. Α, ω-dicarboxylic acid accumulation by acyl-coa oxidase deficient mutants of yarrowia lipolytica. Biotechnol. Lett. 2005, 27, 859–864. [Google Scholar] [CrossRef] [PubMed]
- Wache, Y. Yarrowia lipolytica biotechnological applications: Production of dicarboxylic acids and flagrances by yarrowia lipolytica. Microbiol. Monogr. 2013, 25, 151–170. [Google Scholar]
- Nicaud, J.-M.; Thevenieau, F.; Le Dall, M.-T.; Marchal, R. Production of Dicarboxylic Acids by Improved Mutant Strains of Yarrowia Lipolytica. U.S. Patent 20100041115 A1, 18 February 2010. [Google Scholar]
- Foo, J.L.; Susanto, A.V.; Keasling, J.D.; Leong, S.S.J.; Chang, M.W. Whole-cell biocatalytic and de novo production of alkanes from free fatty acids in saccharomyces cerevisiae. Biotechnol. Bioeng. 2017, 114, 232–237. [Google Scholar] [CrossRef] [PubMed]
- Ledesma-Amaro, R.; Dulermo, R.; Niehus, X.; Nicaud, J.-M. Combining metabolic engineering and process optimization to improve production and secretion of fatty acids. Metab. Eng. 2016, 38, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Cintolesi, A.; Rodríguez-Moyá, M.; Gonzalez, R. Fatty acid oxidation: Systems analysis and applications. Wiley Interdiscip. Rev. Syst. Biol. Med. 2013, 5, 575–585. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, E.; Köhler, V.; Flitsch, S.L.; Turner, N.J. Cytochromes p450 as useful biocatalysts: Addressing the limitations. Chem. Commun. 2011, 47, 2490–2501. [Google Scholar] [CrossRef] [PubMed]
- Endoh-Yamagami, S.; Hirakawa, K.; Morioka, D.; Fukuda, R.; Ohta, A. Basic helix-loop-helix transcription factor heterocomplex of yas1p and yas2p regulates cytochrome p450 expression in response to alkanes in the yeast yarrowia lipolytica. Eukaryot. Cell 2007, 6, 734–743. [Google Scholar] [CrossRef] [PubMed]
- Beopoulos, A.; Mrozova, Z.; Thevenieau, F.; Le Dall, M.T.; Hapala, I.; Papanikolaou, S.; Chardot, T.; Nicaud, J.M. Control of lipid accumulation in the yeast yarrowia lipolytica. Appl. Environ. Microbiol. 2008, 74, 7779–7789. [Google Scholar] [CrossRef] [PubMed]
- Goldbach, V.; Roesle, P.; Mecking, S. Catalytic isomerizing ω-functionalization of fatty acids. ACS Catal. 2015, 5, 5951–5972. [Google Scholar] [CrossRef]
- Isikgor, F.; Becer, C.R. Lignocellulosic biomass: A sustainable platform for production of bio-based chemicals and polymers. Polym. Chem. 2015, 6, 4497–4559. [Google Scholar] [CrossRef]
- Groenewald, M.; Boekhout, T.; Neuveglise, C.; Gaillardin, C.; Dijck, P.W.; Wyss, M. Yarrowia lipolytica: Safety assessment of an oleaginous yeast with a great industrial potential. Crit. Rev. Microbiol. 2014, 40, 187–206. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Coldspring-Harbour Laboratory Press: Plymouth, UK, 2001. [Google Scholar]
- Tan, M.-J.; Chen, X.; Wang, Y.-K.; Liu, G.-L.; Chi, Z.-M. Enhanced citric acid production by a yeast yarrowia lipolytica over-expressing a pyruvate carboxylase gene. Bioprocess Biosyst. Eng. 2016, 39, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.; Jackson, E.N.; Zhu, Q. Sustainable source of omega-3 eicosapentaenoic acid from metabolically engineered yarrowia lipolytica: From fundamental research to commercial production. Appl. Microbiol. Biotechnol. 2015, 99, 1599–1610. [Google Scholar] [CrossRef] [PubMed]
- Chuang, L.-T.; Chen, D.-C.; Nicaud, J.-M.; Madzak, C.; Chen, Y.-H.; Huang, Y.-S. Co-expression of heterologous desaturase genes in yarrowia lipolytica. New Biotechnol. 2010, 27, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Madzak, C.; Nicaud, J.M.; Gaillardin, C. Yarrowia lipolytica. In Production of Recombinant Proteins: Novel Microbial and Eukaryotic Expression Systems; Gellissen, G., Ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2005; pp. 163–189. [Google Scholar]
- Madzak, C. Yarrowia lipolytica: Recent achievements in heterologous protein expression and pathway engineering. Appl. Microbiol. Biotechnol. 2015, 99, 4559–4577. [Google Scholar] [CrossRef] [PubMed]
- Fickers, P.; Le Dall, M.T.; Gaillardin, C.; Thonart, P.; Nicaud, J.M. New disruption cassettes for rapid gene disruption and marker rescue in the yeast yarrowia lipolytica. J. Microbiol. Methods 2003, 55, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Lõoke, M.; Kristjuhan, K.; Kristjuhan, A. Extraction of genomic DNA from yeasts for pcr-based applications. Biotechniques 2011, 50, 325–328. [Google Scholar] [CrossRef] [PubMed]
- O’fallon, J.; Busboom, J.; Nelson, M.; Gaskins, C. A direct method for fatty acid methyl ester synthesis: Application to wet meat tissues, oils, and feedstuffs. J. Anim. Sci. 2007, 85, 1511–1521. [Google Scholar] [CrossRef] [PubMed]
- Abghari, A.; Chen, S. Engineering yarrowia lipolytica for enhanced production of lipid and citric acid. Fermentation 2017, 3, 34. [Google Scholar] [CrossRef]
- Tai, M.; Stephanopoulos, G. Engineering the push and pull of lipid biosynthesis in oleaginous yeast yarrowia lipolytica for biofuel production. Metab. Eng. 2013, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Morin, N.; Cescut, J.; Beopoulos, A.; Lelandais, G.; Le Berre, V.; Uribelarrea, J.-L.; Molina-Jouve, C.; Nicaud, J.-M. Transcriptomic analyses during the transition from biomass production to lipid accumulation in the oleaginous yeast yarrowia lipolytica. PLoS ONE 2011, 6, e27966. [Google Scholar] [CrossRef] [PubMed]
- Kirtz, M.; Klebensberger, J.; Otte, K.B.; Richter, S.M.; Hauer, B. Production of ω-hydroxy octanoic acid with escherichia coli. J. Biotechnol. 2016, 230, 30–33. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Cheng, T.; Zhao, G.; Niu, W.; Guo, J.; Xian, M.; Liu, H. Metabolic engineering of Escherichia coli for the production of hydroxy fatty acids from glucose. BMC Biotechnol. 2016, 16, 26. [Google Scholar] [CrossRef] [PubMed]
- Clomburg, J.M.; Blankschien, M.D.; Vick, J.E.; Chou, A.; Kim, S.; Gonzalez, R. Integrated engineering of β-oxidation reversal and ω-oxidation pathways for the synthesis of medium chain ω-functionalized carboxylic acids. Metab. Eng. 2015, 28, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, L.; Zheng, Y.; Zou, H.; Cao, Y.; Liu, H.; Liu, W.; Xian, M. Biosynthesis of long chain hydroxyfatty acids from glucose by engineered escherichia coli. Bioresour. Technol. 2012, 114, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Picataggio, S.; Rohrer, T.; Eirich, L.D. Method for Increasing the Omega-Hydroxylase Activity in Candida Tropicalis. U.S. Patent 5620878 A, 15 April 1997. [Google Scholar]
- Sumita, T.; Iida, T.; Yamagami, S.; Horiuchi, H.; Takagi, M.; Ohta, A. Ylalk1 encoding the cytochrome p450alk1 in yarrowia lipolytica is transcriptionally induced by n-alkane through two distinct cis-elements on its promoter. Biochem. Biophys. Res. Commun. 2002, 294, 1071–1078. [Google Scholar] [CrossRef]
- Titorenko, V.I.; Rachubinski, R.A. Dynamics of peroxisome assembly and function. Trends Cell Biol. 2001, 11, 22–29. [Google Scholar] [CrossRef]
- Runguphan, W.; Keasling, J.D. Metabolic engineering of saccharomyces cerevisiae for production of fatty acid-derived biofuels and chemicals. Metab. Eng. 2014, 21, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Seip, J.; Jackson, R.; He, H.; Zhu, Q.; Hong, S.-P. Snf1 is a regulator of lipid accumulation in yarrowia lipolytica. Appl. Environ. Microbiol. 2013, 79, 7360–7370. [Google Scholar] [CrossRef] [PubMed]
- Hofbauer, H.F.; Schopf, F.H.; Schleifer, H.; Knittelfelder, O.L.; Pieber, B.; Rechberger, G.N.; Wolinski, H.; Gaspar, M.L.; Kappe, C.O.; Stadlmann, J. Regulation of gene expression through a transcriptional repressor that senses acyl-chain length in membrane phospholipids. Dev. Cell 2014, 29, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Thevenieau, F.; Le Dall, M.T.; Nthangeni, B.; Mauersberger, S.; Marchal, R.; Nicaud, J.M. Characterization of yarrowia lipolytica mutants affected in hydrophobic substrate utilization. Fungal Genet. Biol. 2007, 44, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Scharnewski, M.; Pongdontri, P.; Mora, G.; Hoppert, M.; Fulda, M. Mutants of saccharomyces cerevisiae deficient in acyl-coa synthetases secrete fatty acids due to interrupted fatty acid recycling. FEBS J. 2008, 275, 2765–2778. [Google Scholar] [CrossRef] [PubMed]
- Dulermo, R.; Gamboa-Meléndez, H.; Ledesma-Amaro, R.; Thévenieau, F.; Nicaud, J.-M. Unraveling fatty acid transport and activation mechanisms in yarrowia lipolytica. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2015, 1851, 1202–1217. [Google Scholar] [CrossRef] [PubMed]
- Dulermo, R.; Gamboa-Meléndez, H.; Ledesma, R.; Thevenieau, F.; Nicaud, J.-M. Yarrowia lipolytica aal genes are involved in peroxisomal fatty acid activation. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2016, 1861, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, R.; Ohta, A. Utilization of hydrophobic substrate by yarrowia lipolytica. In Yarrowia Lipolytica Genetics, Genomics, and Physiology; Barth, G., Ed.; Springer: Berlin/Heidelberg, Gemany, 2013; pp. 111–119. [Google Scholar]
- Park, J.S.; Iwama, R.; Kobayashi, S.; Ohta, A.; Horiuchi, H.; Fukuda, R. Involvement of acyl-coa synthetase genes in n-alkane assimilation and fatty acid utilization in yeast yarrowia lipolytica. FEMS Yeast Res. 2015, 15, fov031. [Google Scholar] [CrossRef]
- Krivoruchko, A.; Zhang, Y.; Siewers, V.; Chen, Y.; Nielsen, J. Microbial acetyl-coa metabolism and metabolic engineering. Metab. Eng. 2015, 28, 28–42. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, J.; Lee, J.; Chen, W.N. Enhancement of free fatty acid production in saccharomyces cerevisiae by control of fatty acyl-coa metabolism. Appl. Microbiol. Biotechnol. 2014, 98, 6739–6750. [Google Scholar] [CrossRef] [PubMed]
- Petschnigg, J.; Wolinski, H.; Kolb, D.; Zellnig, G.; Kurat, C.F.; Natter, K.; Kohlwein, S.D. Good fat, essential cellular requirements for triacylglycerol synthesis to maintain membrane homeostasis in yeast. J. Biol. Chem. 2009, 284, 30981–30993. [Google Scholar] [CrossRef] [PubMed]
- Stefan, A.; Ugolini, L.; Lazzeri, L.; Conte, E.; Hochkoeppler, A. The expression of the cuphea palustris thioesterase cpfatb2 in yarrowia lipolytica triggers oleic acid accumulation. Biotechnol. Prog. 2015, 32, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Qiao, K.; Stephanopoulos, G. Engineering oxidative stress defense pathways to build a robust lipid production platform in yarrowia lipolytica. Biotechnol. Bioeng. 2017, 114, 1521–1530. [Google Scholar] [CrossRef] [PubMed]
- Kohlwein, S.D. Triacylglycerol homeostasis: Insights from yeast. J. Biol. Chem. 2010, 285, 15663–15667. [Google Scholar] [CrossRef] [PubMed]
- Hardwick, J.P. Cytochrome p450 omega hydroxylase (cyp4) function in fatty acid metabolism and metabolic diseases. Biochem. Pharmacol. 2008, 75, 2263–2275. [Google Scholar] [CrossRef] [PubMed]
- Björkhem, I. On the mechanism of regulation of omega oxidation of fatty acids. J. Biol. Chem. 1976, 251, 5259–5266. [Google Scholar] [PubMed]
- Wanders, R.J.; Komen, J.; Kemp, S. Fatty acid omega-oxidation as a rescue pathway for fatty acid oxidation disorders in humans. FEBS J. 2011, 278, 182–194. [Google Scholar] [CrossRef] [PubMed]
- Geier, M.; Braun, A.; Emmerstorfer, A.; Pichler, H.; Glieder, A. Production of human cytochrome p450 2d6 drug metabolites with recombinant microbes–a comparative study. Biotechnol. J. 2012, 7, 1346–1358. [Google Scholar] [CrossRef] [PubMed]
- Eirich, L.D.; Craft, D.L.; Steinberg, L.; Asif, A.; Eschenfeldt, W.H.; Stols, L.; Donnelly, M.I.; Wilson, C.R. Cloning and characterization of three fatty alcohol oxidase genes from candida tropicalis strain atcc 20336. Appl. Environ. Microbiol. 2004, 70, 4872–4879. [Google Scholar] [CrossRef] [PubMed]
- Kang, W.-R.; Seo, M.-J.; An, J.-U.; Shin, K.-C.; Oh, D.-K. Production of δ-decalactone from linoleic acid via 13-hydroxy-9 (z)-octadecenoic acid intermediate by one-pot reaction using linoleate 13-hydratase and whole yarrowia lipolytica cells. Biotechnol. Lett. 2016, 38, 817–823. [Google Scholar] [CrossRef] [PubMed]
- Iwama, R.; Kobayashi, S.; Ohta, A.; Horiuchi, H.; Fukuda, R. Fatty aldehyde dehydrogenase multigene family involved in the assimilation of n-alkanes in yarrowia lipolytica. J. Biol. Chem. 2014, 289, 33275–33286. [Google Scholar] [CrossRef] [PubMed]
- Gatter, M.; Matthaus, F.; Barth, G. Yeast Strains and Method for the Production of Omega-Hydroxy Fatty Acids and Dicarboxylic Acids. U.S. Patent 20160304913 A1, 20 October 2016. [Google Scholar]
- Vanhanen, S.; West, M.; Kroon, J.T.; Lindner, N.; Casey, J.; Cheng, Q.; Elborough, K.M.; Slabas, A.R. A consensus sequence for long-chain fatty-acid alcohol oxidases from candida identifies a family of genes involved in lipid ω-oxidation in yeast with homologues in plants and bacteria. J. Biol. Chem. 2000, 275, 4445–4452. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, C.; Fang, X.; Cao, Z.A. Optimal ph control strategy for high-level production of long-chain α, ω-dicarboxylic acid by candida tropicalis. Enzym. Microb. Technol. 2004, 34, 73–77. [Google Scholar] [CrossRef]
- Lin, R.; Cao, Z.; Zhu, T.; Zhang, Z. Secretion in long-chain dicarboxylic acid fermentation. Bioprocess Eng. 2000, 22, 391–396. [Google Scholar] [CrossRef]
- Bankar, A.V.; Kumar, A.R.; Zinjarde, S.S. Environmental and industrial applications of yarrowia lipolytica. Appl. Microbiol. Biotechnol. 2009, 84, 847–865. [Google Scholar] [CrossRef] [PubMed]
- Casal, M.; Paiva, S.; Queirós, O.; Soares-Silva, I. Transport of carboxylic acids in yeasts. FEMS Microbiol. Rev. 2008, 32, 974–994. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, C.; Xie, L.; Cao, Z.A. Intracellular ph and metabolic activity of long-chain dicar☐ ylic acid-producing yeastCandida tropicalis. J. Biosci. Bioeng. 2003, 96, 349–353. [Google Scholar] [CrossRef]
- Kamisaka, Y.; Tomita, N.; Kimura, K.; Kainou, K.; Uemura, H. DGA1 (diacylglycerol acyltransferase gene) overexpression and leucine biosynthesis significantly increase lipid accumulation in the Δsnf2 disruptant of saccharomyces cerevisiae. Biochem. J. 2007, 408, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Lundemo, M.T.; Woodley, J.M. Guidelines for development and implementation of biocatalytic p450 processes. Appl. Microbiol. Biotechnol. 2015, 99, 2465–2483. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhao, F.; Chen, D.; Li, D. Biotransformation of phytosterol to produce androsta-diene-dione by resting cells of mycobacterium in cloud point system. Process Biochem. 2006, 41, 557–561. [Google Scholar] [CrossRef]
- Vandenberghe, L.P.S.; Soccol, C.R.; Pandey, A.; Lebeault, J.-M. Microbial production of citric acid. Braz. Arch. Biol. Technol. 1999, 42, 263–276. [Google Scholar] [CrossRef]
- Papanikolaou, S.; Aggelis, G. Selective uptake of fatty acids by the yeast yarrowia lipolytica. Eur. J. Lipid Sci. Technol. 2003, 105, 651–655. [Google Scholar] [CrossRef]
- Hirakawa, K.; Kobayashi, S.; Inoue, T.; Endoh-Yamagami, S.; Fukuda, R.; Ohta, A. Yas3p, an opi1 family transcription factor, regulates cytochrome p450 expression in response to n-alkanes in yarrowia lipolytica. J. Biol. Chem. 2009, 284, 7126–7137. [Google Scholar] [CrossRef] [PubMed]
- Beopoulos, A.; Haddouche, R.; Kabran, P.; Dulermo, T.; Chardot, T.; Nicaud, J.-M. Identification and characterization of dga2, an acyltransferase of the dgat1 acyl-coa: Diacylglycerol acyltransferase family in the oleaginous yeast yarrowia lipolytica. New insights into the storage lipid metabolism of oleaginous yeasts. Appl. Microbiol. Biotechnol. 2012, 93, 1523–1537. [Google Scholar] [CrossRef] [PubMed]
- Wasylenko, T.M.; Ahn, W.S.; Stephanopoulos, G. The oxidative pentose phosphate pathway is the primary source of nadph for lipid overproduction from glucose in yarrowia lipolytica. Metab. Eng. 2015, 30, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Silverman, A.M.; Qiao, K.; Xu, P.; Stephanopoulos, G. Functional overexpression and characterization of lipogenesis-related genes in the oleaginous yeast yarrowia lipolytica. Appl. Microbiol. Biotechnol. 2016, 100, 3781–3798. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wu, C.; Wu, Q.; Dai, J.; Song, Y. Metabolic flux analysis of lipid biosynthesis in the yeast yarrowia lipolytica using 13 c-labled glucose and gas chromatography-mass spectrometry. PLoS ONE 2016, 11, e0159187. [Google Scholar] [CrossRef]
- Fang, F.; Dai, B.; Zhao, G.; Zhao, H.; Sun, C.; Liu, H.; Xian, M. In depth understanding the molecular response to the enhanced secretion of fatty acids in saccharomyces cerevisiae due to one-step gene deletion of acyl-coa synthetases. Process Biochem. 2016, 51, 1162–1174. [Google Scholar] [CrossRef]
- Jin, Z.; Wong, A.; Foo, J.L.; Ng, J.; Cao, Y.X.; Chang, M.W.; Yuan, Y.J. Engineering saccharomyces cerevisiae to produce odd chain-length fatty alcohols. Biotechnol. Bioeng. 2016, 113, 842–851. [Google Scholar] [CrossRef] [PubMed]
- Vohra, A.; Syal, P.; Madan, A. Probiotic yeasts in livestock sector. Anim. Feed Sci. Technol. 2016, 219, 31–47. [Google Scholar] [CrossRef]
- Martinez, C.A.; Rupashinghe, S.G. Cytochrome p450 bioreactors in the pharmaceutical industry: Challenges and opportunities. Curr. Top. Med. Chem. 2013, 13, 1470–1490. [Google Scholar] [CrossRef] [PubMed]
- Holtmann, D.; Hollmann, F. The oxygen dilemma: A severe challenge for the application of monooxygenases? ChemBioChem 2016, 17, 1391–1398. [Google Scholar] [CrossRef] [PubMed]
- Grand View Research, Inc. Long Chain Dicarboxylic Acid Market Analysis; Grand View Research: Maharashtra, India, 2016. [Google Scholar]
- Stempfle, F.; Ortmann, P.; Mecking, S. Long-chain aliphatic polymers to bridge the gap between semicrystalline polyolefins and traditional polycondensates. Chem. Rev. 2016, 116, 4597–4641. [Google Scholar] [CrossRef] [PubMed]
- De Montellano, P.R.O. Cytochrome p450: Structure, Mechanism, and Biochemistry; Springer Science & Business Media: Berlin, Gemany, 2005. [Google Scholar]
- Cambon, E.; Piamtongkam, R.; Bordes, F.; Duquesne, S.; Laguerre, S.; Nicaud, J.-M.; Marty, A. A new yarrowia lipolytica expression system: An efficient tool for rapid and reliable kinetic analysis of improved enzymes. Enzym. Microb. Technol. 2010, 47, 91–96. [Google Scholar] [CrossRef]
- Xu, P.; Gu, Q.; Wang, W.; Wong, L.; Bower, A.G.W.; Collins, C.H.; Koffas, M.A.G. Modular optimization of multi-gene pathways for fatty acids production in E. coli. Nat. Commun. 2013, 4, 1409. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, P.G.; Ferreira, R.; Zhou, Y.J.; Siewers, V.; Nielsen, J. Dynamic regulation of fatty acid pools for improved production of fatty alcohols in saccharomyces cerevisiae. Microb. Cell Factories 2017, 16, 45. [Google Scholar] [CrossRef] [PubMed]
- Dulermo, T.; Tréton, B.; Beopoulos, A.; Gnankon, A.P.K.; Haddouche, R.; Nicaud, J.-M. Characterization of the two intracellular lipases of y. Lipolytica encoded by tgl3 and tgl4 genes: New insights into the role of intracellular lipases and lipid body organisation. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2013, 1831, 1486–1495. [Google Scholar] [CrossRef] [PubMed]
- Sagehashi, Y.; Horiuchi, H.; Fukuda, R.; Ohta, A. Identification and characterization of a gene encoding an abc transporter expressed in the dicarboxylic acid-producing yeast candida maltosa. Biosci. Biotechnol. Biochem. 2013, 77, 2502–2504. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Ling, H.; Chang, M.W. Transporter engineering for improved tolerance against alkane biofuels in saccharomyces cerevisiae. Biotechnol. Biofuels 2013, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Kavšček, M.; Bhutada, G.; Madl, T.; Natter, K. Optimization of lipid production with a genome-scale model of yarrowia lipolytica. BMC Syst. Biol. 2015, 9, 72. [Google Scholar] [CrossRef] [PubMed]
- Cescut, J. Accumulation D’acylglycérols par des Espèces Levuriennes à Usage Carburant Aéronautique: Physiologie et Performances de Procédés; INSA Toulouse: Toulouse, France, 2009; (In French). Available online: http://www.theses.fr/2009ISAT0022 (accessed on 22 July 2017).
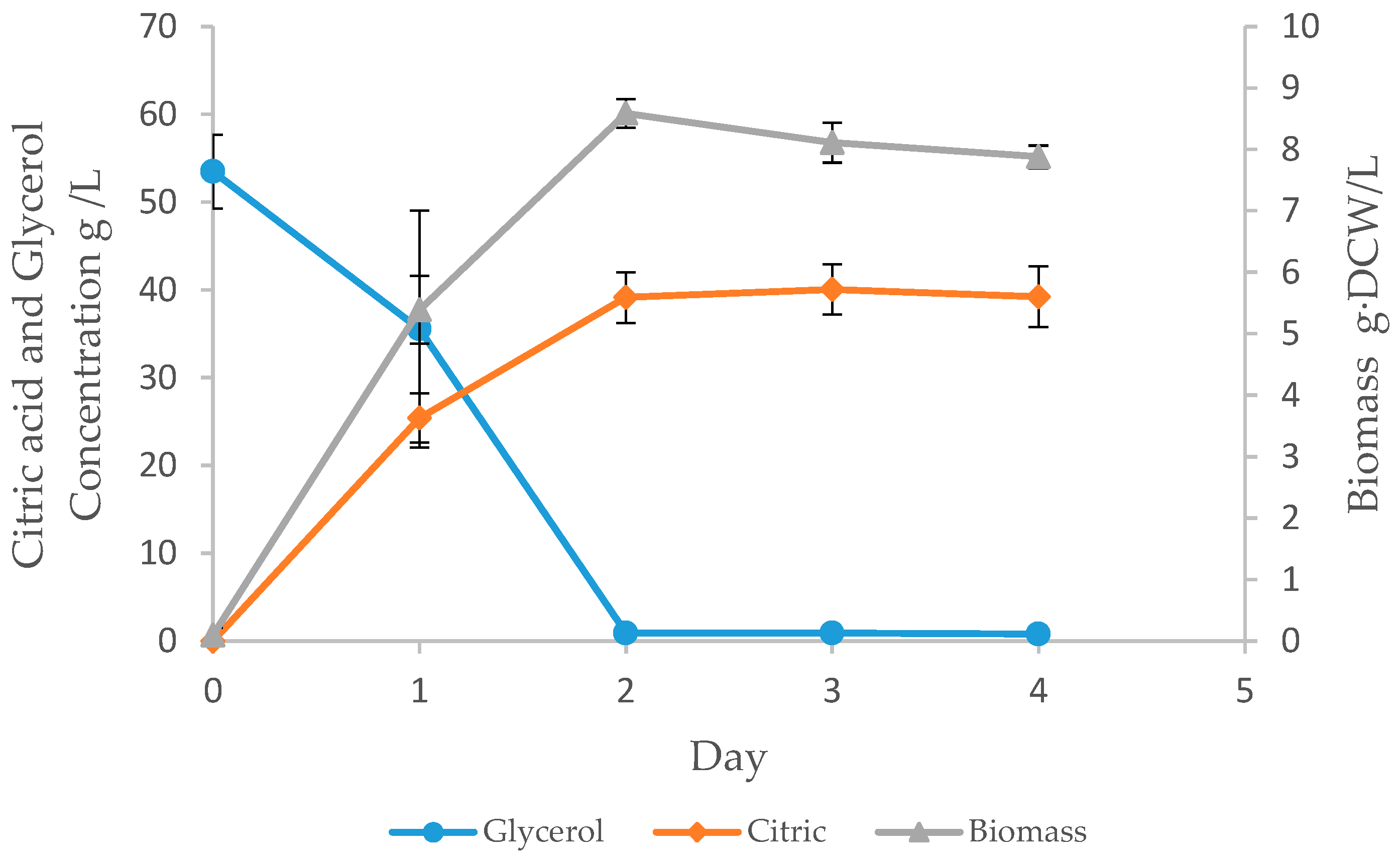


| Y. lipolytica Strains Names | Strain Genotypes | Gene Configurations | Reference |
|---|---|---|---|
| H222 (W) [wild type] | MatA | [42] | |
| H222ΔP (HP-U) [leu+, ura−] | MatA ura3-302::SUC2 Δpox1 Δpox2 Δpox3 Δpox4 Δpox5 Δpox6 | [42] | |
| H222ΔPΔLΔSΔF (F) [leu−, ura+] | Same as HP-U, +Δleu2 Δsnf1 Δfaa1 Δsnf1::URA3 | loxR-URA3-loxP flanked by SNF1 upstream and downstream | This study |
| H222ΔPΔLΔSΔF ΔL +ALK5 YlCPR YlFAO1 (P) [leu−, ura+] | Same as F, +Δleu2::URA3 YlALK5 YlCPR YFAO1 | loxR-URA3-loxP flanked by SNF1 upstream and downstream | This study |
| H222ΔPΔLΔSΔF ΔL +ALK5 YlCPR YlFAO1 ++ALK5 (M) [leu−, ura+] | Same as P, +YlALK5 | Multiple-copy integration of YlALK5 using zeta based integrative vector pINA1291(this strain was selected after screening of 10–20 transformants for their growth and production capacities) | This study |
| Vector Names | Map | Features |
|---|---|---|
| Cre (CR) | 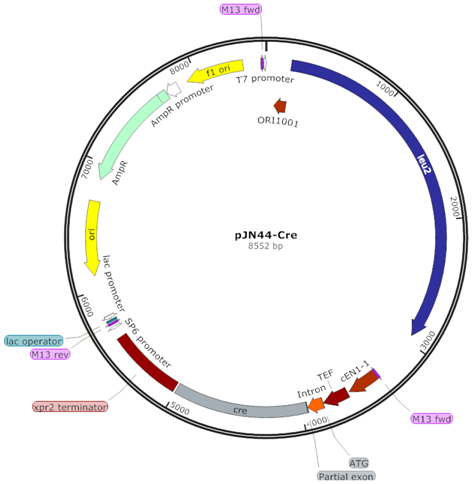 | Cre recombinase flanked by TEFin promoter and Xpr2 terminator |
| pGR12 YlALK5 (5) |  | Centromeric (CEN) replicative vector, low-copy CEN plasmids (1–2 copies/cell~1.6 plasmid copies/cell), YlALK5 controlled by PFBA-Tlip1, leucine selection marker |
| pGR51 YlCPR (C) | 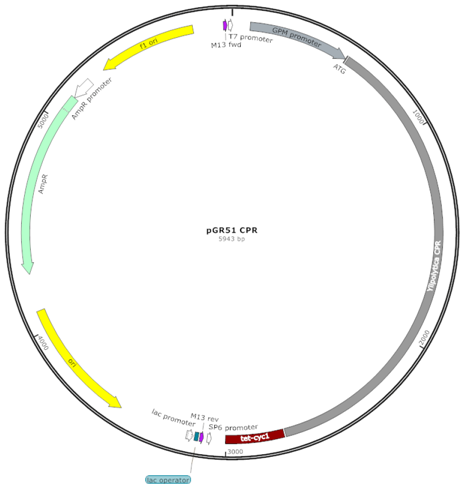 | Centromeric (CEN) replicative vector, YlCPR controlled by PGPM-Tcyc1 |
| pJN44 YlFAO (F) | 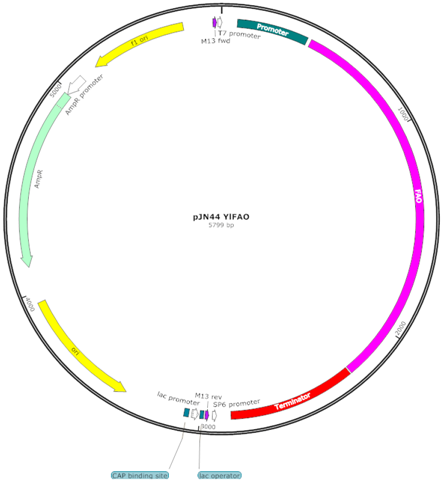 | Centromeric (CEN) replicative vector, YlFAO1 controlled by PTEFin-Txpr2 |
| ALK5 CPR FAO (5CF) |  | CEN replicative vector, PFBA-YlALK5 Tlip1 PGPM-YlCPR Tcyc1 PTEFin-YlFAO1 Txpr2 |
| Leu 5cf (L5CF) |  | Uracil selection marker flanked by LEU2 upstream and PFBA-YlALK5 Tlip1 PGPM-YlCPR Tcyc1 PTEFin-YlFAO1 Txpr2 LEU2 downstream sequences |
| pINA1291 ALK5 (Z5) | 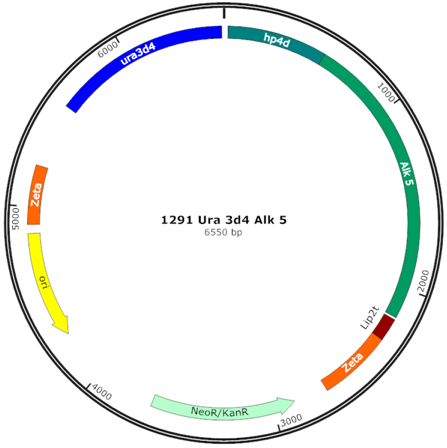 | Integrative vector, hp4d- YlALK5 Tlip2, ura3d4 defective marker, zeta region for multi-copy integration |
| LEU (LU) | 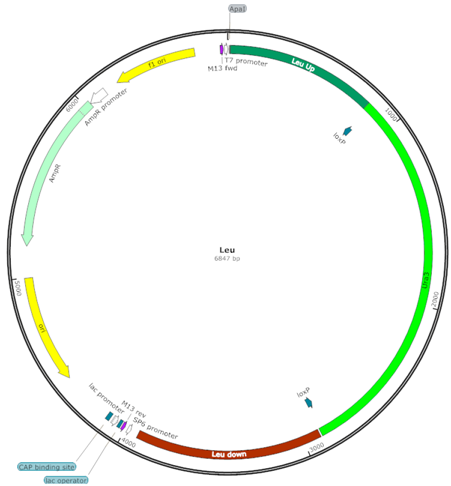 | Uracil selection marker flanked by LEU2 upstream and downstream sequences |
| SNF (SU) | 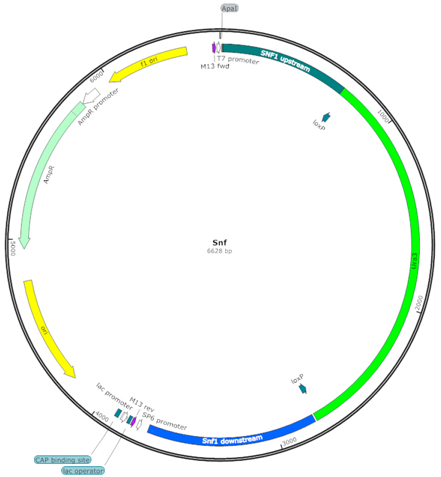 | Uracil selection marker flanked by SNF1 upstream and downstream sequences |
| FAA (FU) |  | Uracil selection marker flanked by FAA1 upstream and downstream sequences |
| No. | Name | Sequence (5′→›3′, Underlined Restriction Site) |
|---|---|---|
| 1 | Alk 5 F HindIII | GAGCGAAAGCTTATGCTACAACTCTTTGGCGTCC |
| 2 | Alk 5 R PstI | CTTAGA CTGCAG CTACGCCTTCTCACCCTTATACA |
| 3 | Alk 5 Zeta F A | AATGCTACAACTCTTTGGCGTCC |
| 4 | Alk 5 Zeta KpnI R | TTGCAAGGTACCCTACGCCTTCTCACCCTTATACATCT |
| 5 | YlCPR F HindIII | GAGCGAAAGCTT ATGGCTCTACTCGAC TCTC |
| 6 | YlCPR R SmaI | GTTAT CCCGGG CTACCACACATCTTCCTGG |
| 7 | FAO1F NdeI | CCTCA CATATGATGTCTGACGACAAGCACACT |
| 8 | FAO1R SmaI | GTTAT CCCGGG AGGATCTCCGACCTCGAATC |
| 9 | LEU2 up F ApaI | CTATAGGGCCC ACCGGCAAGATCTCGTTAAGACAC |
| 10 | LEU2 up R XbaI | GATCCTCTAGATGTGTGTGGTTGTATGTGTGATGTGG |
| 11 | LEU2 down F SpeI | CTGGACTAGTCTCTATAAAAAGGGCCCAGCCCTG |
| 12 | LEU2 down R NdeI | CCTCACATATG GACAGCCTTGACAACTTGGTTGTTG |
| 13 | LEU2 F Ura | TACAGTTGTAACTATGGTGCTTATCTGGG |
| 14 | LEU2 Ura R | CCTTGGGAACCACCACCGT |
| 15 | LEU2 Ura F | ACTTCCTGGAGGCAGAAGAACTT |
| 16 | LEU2 R Ura | ATAGCAAATTTAGTCGTCGAGAAAGGGTC |
| 17 | SNF1 up F ApaI | CAATTGGGCCCGTGATCAAAGCATGAGATACTGTCAAGG |
| 18 | SNF1 up R XbaI | GATCCTCTAGAGAGGTGGTGGAAGGAGTGGTATGTAGTC |
| 19 | SNF1 down F SpeI | CTGGACTAGT TCATTAATACGTTTCCCTGGTG |
| 20 | SNF1 down R NdeI | CCTCACATATGGGAATTCGTGCAGAAGAACA |
| 21 | SNF1 F Ura | GCGGGAAATCAAGATTGAGA |
| 22 | SNF1Ura R | CGGTCCATTTCTCACCAACT |
| 23 | SNF1 Ura F | CCTGGAGGCAGAAGAACTTG |
| 24 | SNF1 R Ura | ACTACTGGCGGACTTTGTGG |
| 25 | FAA1 up F ApaI | CAATT GGGCCC CCAGGTCTCAGTTGCACTTGC |
| 26 | FAA1 up R XbaI | GATCC TCTAGA CAAATTATACCCCTCATCTCTCTAGGACA |
| 27 | FAA1 down F SpeI | CT GGACTAGTTTGGTGAGCCCACCGC |
| 28 | FAA1 down R NdeI | CCTCACATATGAACCTCCAGCAGACTAACTAGAACA |
| 29 | FAA1 F Ura | ACTGTAGCTAGATGGGTGCC |
| 30 | FAA1 Ura R | CGGTCCATTTCTCACCAACT |
| 31 | FAA1 Ura F | CCTGGAGGCAGAAGAACTTG |
| 32 | FAA1 R Ura | ACCAGCCCAGCCGG |
| 33 | LEU2 up F Ura | TCATGTTCGTGGAGGGGAG |
| 34 | LEU2 up Ura R | AAAACGCAGCTGTCAGACC |
| 35 | LEU2 down F FAO | GGAGTCCAAGCCCTTCGA |
| 36 | LEU2 down R | CACAAGACGTCAACTAAAGCGT |
| DCW (g/L) | Citric Acid (g/L) | LCDCA Titer (g/L) | LCDCA Yield (g/g) | LCDCA Productivity (g/L·h) |
|---|---|---|---|---|
| 8.58 ± 0.23 | 39.2 ± 3.5 | 3.49 ± 0.14 | 0.06 | 0.04 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abghari, A.; Madzak, C.; Chen, S. Combinatorial Engineering of Yarrowia lipolytica as a Promising Cell Biorefinery Platform for the de novo Production of Multi-Purpose Long Chain Dicarboxylic Acids. Fermentation 2017, 3, 40. https://doi.org/10.3390/fermentation3030040
Abghari A, Madzak C, Chen S. Combinatorial Engineering of Yarrowia lipolytica as a Promising Cell Biorefinery Platform for the de novo Production of Multi-Purpose Long Chain Dicarboxylic Acids. Fermentation. 2017; 3(3):40. https://doi.org/10.3390/fermentation3030040
Chicago/Turabian StyleAbghari, Ali, Catherine Madzak, and Shulin Chen. 2017. "Combinatorial Engineering of Yarrowia lipolytica as a Promising Cell Biorefinery Platform for the de novo Production of Multi-Purpose Long Chain Dicarboxylic Acids" Fermentation 3, no. 3: 40. https://doi.org/10.3390/fermentation3030040