Partial Characterization of Digestive Proteases in the Green Cichlid, Cichlasoma beani
Abstract
:1. Introduction
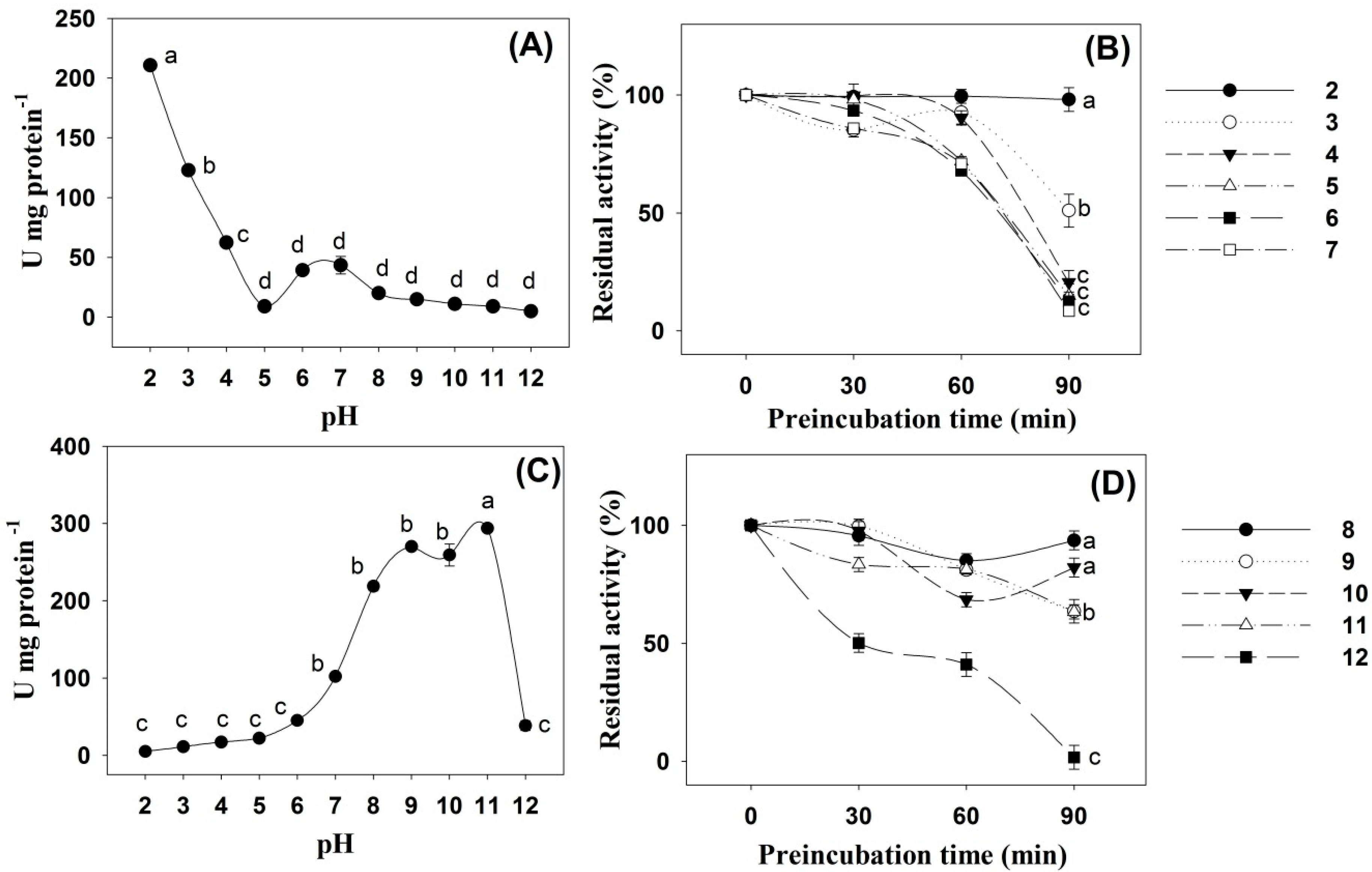
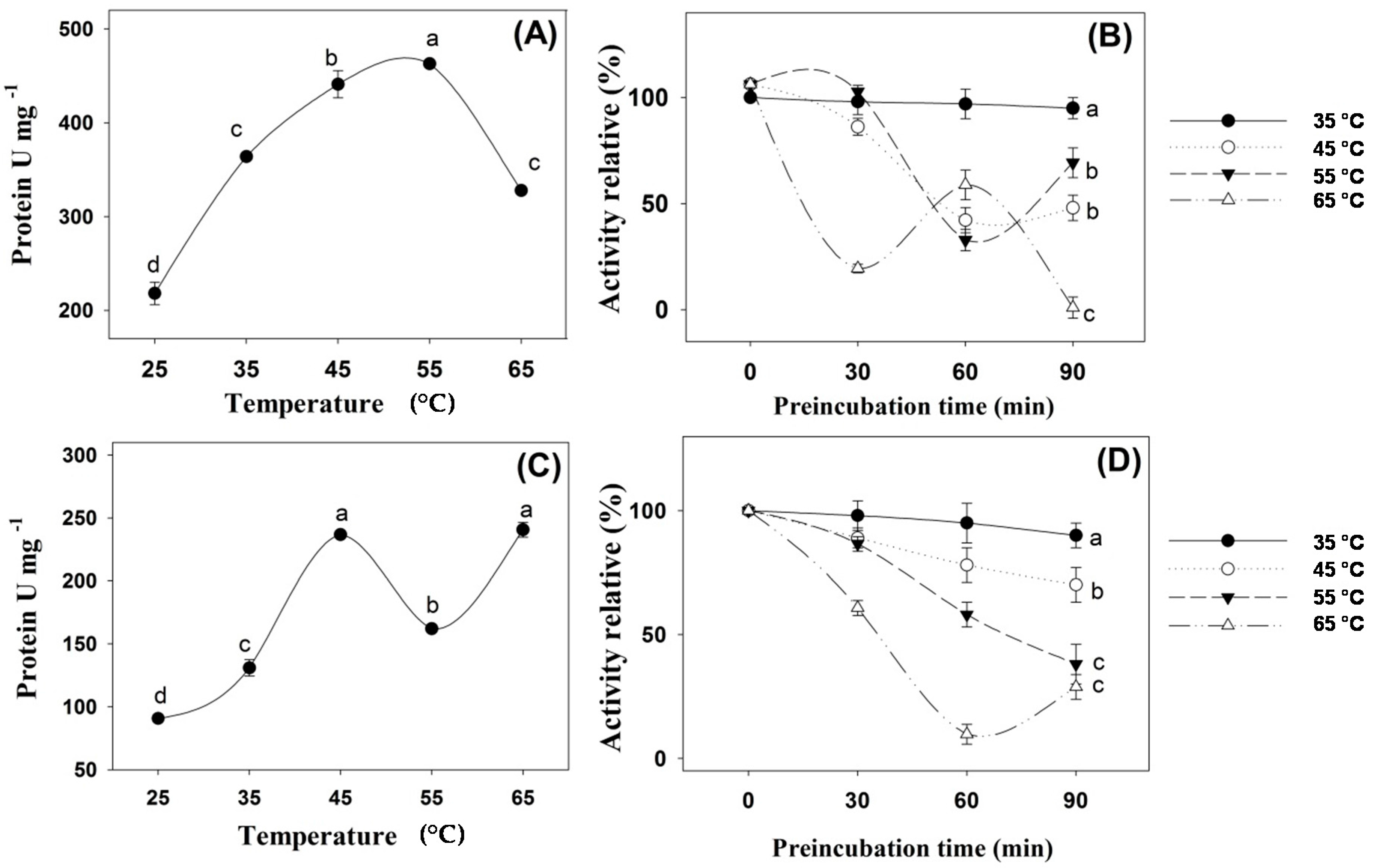
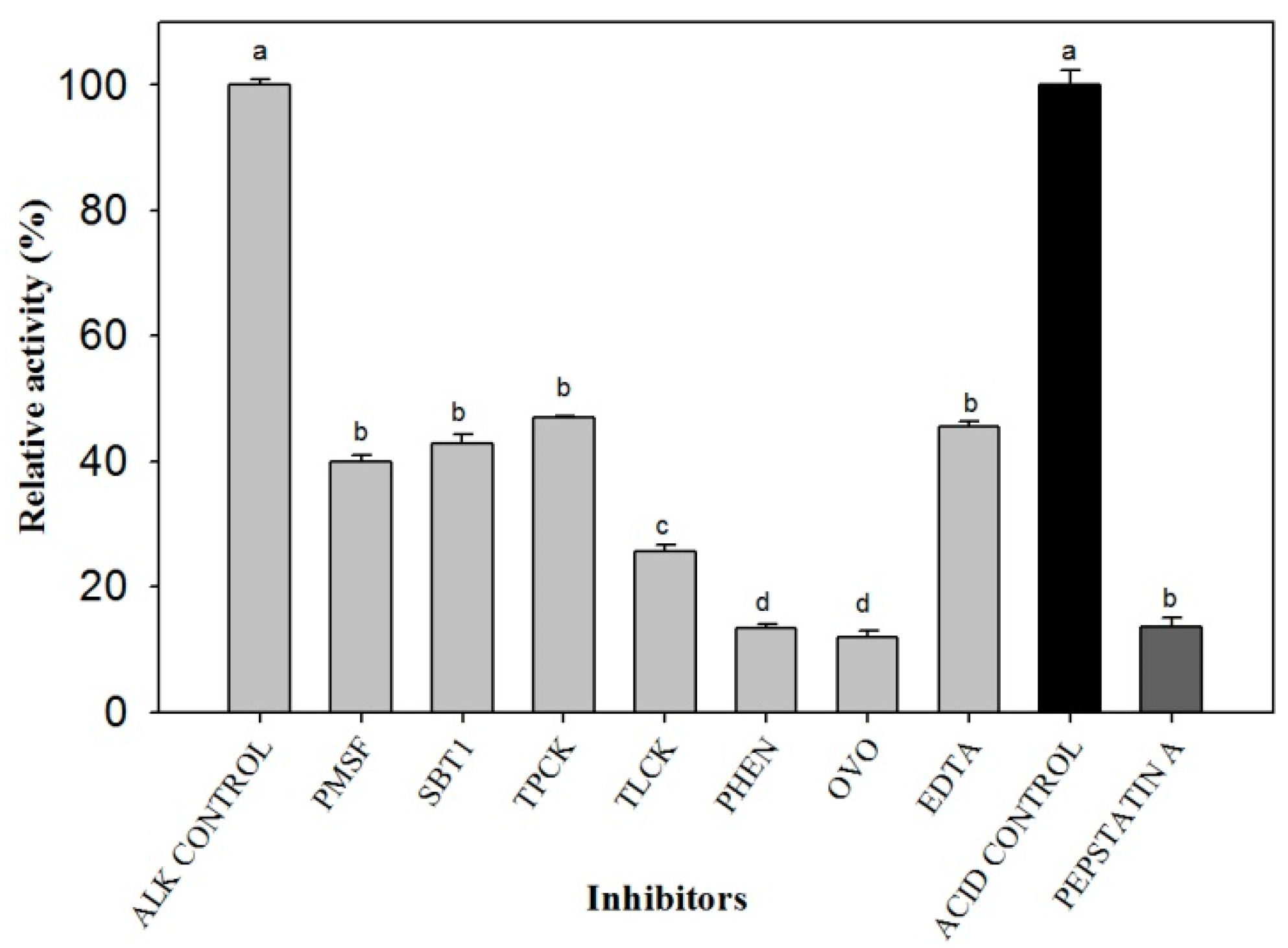
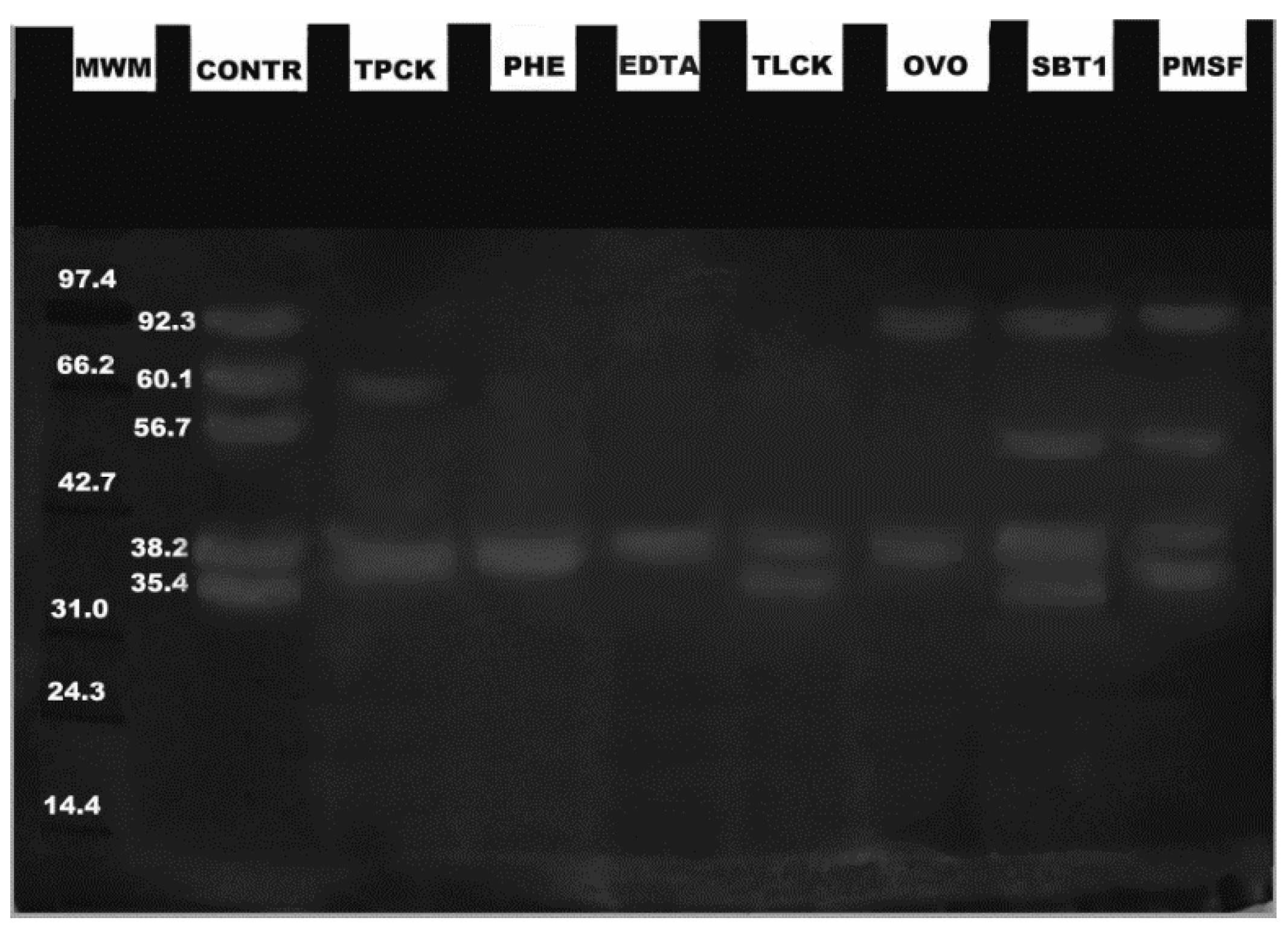
2. Results
3. Discussion
4. Materials and Methods
4.1. Sample Preparation
4.2. Biochemical Analyses
4.3. Effects of pH and Temperature on Digestive Enzyme Activity
4.4. Zymograms and Effects of Inhibitors
4.5. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Baltazar, P.M. La tilapia en el Perú: Acuicultura, mercado y perspectivas. Rev. Peru. Biol. 2007, 13, 267–273. [Google Scholar] [CrossRef]
- Márquez, C.G. Producción por acuicultura sustentable de pegelagarto en Comalcalco, Tabasco. Available online: https://sgp.undp.org/index.php?option=com_sgpprojects&view=projectdetail&id=15073&Itemid=272 (accessed on 28 November 2016).
- García-Lizárraga, M.A.; Soto-Franco, F.E.; Velazco-Arce, J.M.; Velázquez-Abunader, J.I.; Ramírez-Pérez, J.; Peña-Messina, S.E. Population structure and reproductive behavior of Sinaloa cichlid Cichlasoma beani (Jordan, 1889) in a tropical reservoir. Neotrop. Ichthyol. 2011, 9, 593–599. [Google Scholar] [CrossRef]
- Aragón-Flores, E.A.; Valdez-Hernández, E.F.; Martínez-Cárdenas, L.; Castañeda-Chávez, M.R.; Gonzales-Díaz, A.A.; Soria-Barreto, M.; Peña-Messina, E. Effect of Stocking Density on Growth, Survival, and Condition of the Mexican Cichlid Cichlasoma beani. J. World Aquac. Soc. 2014, 45, 447–453. [Google Scholar] [CrossRef]
- Martínez-Cárdenas, L.; Valdez-Hernández, E.F.; González-Díaz, A.A.; Soria-Barreto, M.; Castañeda-Chávez, M.R.; Ruiz-Velazco, J.M.; Robles-Bermúdez, A. Preliminary observations on Cichlasoma beani in culture conditions. Lat. Am. J. Aquat. Res. 2014, 42, 639–643. [Google Scholar] [CrossRef]
- López-Ramírez, G.; Cuenca-Soria, C.A.; Álvarez-González, C.A.; Tovar-Ramírez, D.; Ortiz-Galindo, J.L.; Perales-García, N.; Márquez-Couturier, G.; Arias-Rodríguez, L.; Indy, J.R.; Contreras-Sánchez, W.M.; et al. Development of digestive enzymes in larvae of Mayan cichlid Cichlasoma urophthalmus. Fish Physiol. Biochem. 2011, 37, 197–208. [Google Scholar]
- Cuenca-Soria, C.A.; Álvarez-González, C.A.; Ortiz-Galindo, J.L.; Nolasco-Soria, H.; Tovar-Ramírez, D.; Guerrero-Zárate, R.; Gisbert, E. Partial characterization of digestive proteases of the Mayan cichlid Cichlasoma urophthalmus. Fish Physiol. Biochem. 2014, 40, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Toledo Solís, F.J.; Márquez, G.; Uscanga, A.; Guerrero-Zárate, R.; Perales- García, N.; Martín García, R.; Contreras, W.; Camarillo-Coop, S.; Álvarez-González, C.A. Partial characterization of digestive proteases of the three spot cichlid Cichlasoma trimaculatum (Günter 1867). Aquac. Nutr. 2015, 22, 1230–1238. [Google Scholar] [CrossRef]
- Kolkovski, S. Digestive enzymes in fish larvae and juveniles-implications and applications to formulated diets. Aquaculture 2001, 200, 181–201. [Google Scholar] [CrossRef]
- Lazo, J.P.; Mendoza, R.; Holt, G.J.; Aguilera, C.; Arnold, C.R. Characterization of digestive enzymes during larval development of red drum (Sciaenops ocellatus). Aquaculture 2007, 265, 194–205. [Google Scholar] [CrossRef]
- Jonas, E.; Ragyanski, M.; Olah, J.; Borros, L. Proteolytic digestive enzymes of carnivorous (Silurus glanis, L.), herbivorous (Hypophthalmichthys molitrix, VAL) and omnivorous (Cyprinus carpio, L.) fishes. Aquaculture 1983, 30, 145–154. [Google Scholar] [CrossRef]
- Moyano, F.J. Bioquímica Digestiva en Especies Acuicultivadas: Aplicaciones en Nutrición. In Proceedings of the VIII Simposium Internacional de Nutrición Acuícola, Universidad Autónoma de Nuevo León, Monterrey, Nuevo León, México, 15–17 November 2006; Cruz, S.E., Ricque, M.R., Tapia, S.M., Nieto, L.M., Villareal, C.D., Puello, C.A., García, O.A., Goytortua-Bores, E., Eds.; UANL: Monterrey, Nuevo León, México, 2006. [Google Scholar]
- Álvarez-González, C.A.; Márquez-Couturier, G.; Arias-Rodríguez, L.; Contreras-Sánchez, W.M.; Uscanga-Martínez, A.; Perales-García, N.; Moyano-López, F.J.; Hernández-Jiménez, R.; Civera-Cerecedo, R.; et al. Avances en la Fisiología Digestiva y Nutrición de la Mojarra Tenguayaca Petenia splendida. In Proceedings of the Memoria del IX Avances en Nutrición Acuícola y IX Simposio Internacional de Nutrición Acuícola, Universidad Autónoma de Nuevo León, Monterrey, Nuevo León, México, 24–27 November 2008; Cruz, S.E., Ricque, M.D., Tapia, S.M., Nieto, L.M., Villareal, C.D., Lazo, J.P., Viana, T.V., Eds.; UANL: Monterrey, Nuevo León, México, 2008; pp. 135–235. [Google Scholar]
- Salvensen, G.; Nagase, H. Inhibition of proteolytic enzymes. In Proteolytic Enzymes; Beynon, R., Bond, J.S., Eds.; IRL Press: Oxford, UK, 1989; pp. 83–104. [Google Scholar]
- García-Carreño, F.L.; Dimes, L.E.; Haard, N.F. Substrate-gel electrophoresis for composition and molecular weight of proteinases or proteinaceous proteinase inhibitors. Anal. Biochem. 1993, 214, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-González, C.A.; Gabriela-Cortés, G.; Jiménez-Martínez, L.D.; Sánchez-Zamora, A.; Arena-Ortiz, G.; Martínez-Bruguete, T.; Tovar-Ramírez, D.; Concha-Frías, B.; Márquez-Couturier, G.; Perales-García, N.; et al. Avances en la fisiología digestiva del robalo blanco (Centropomus undecimalis) en Tabasco, México. In Proceedings of the Memorias del X Avances en Nutrición Acuícola y X Simposio Internacional de Nutrición Acuícola, Universidad Autónoma de Nuevo León, Monterrey, México, 8–10 November 2010; Cruz-Suarez, L.E., Ricque-Marie, D., Tapia-Salazar, M., Nieto-López, M.G., Villarreal-Cavazos, D.A., Eds.; Gamboa-Delgado: Monterrey, Nuevo León, México, 2010; pp. 99–232. [Google Scholar]
- Essed, Z.; Fernández, I.; Alarcón, F.J.; Moyano, F.J. Caracterización de la actividad proteasa digestiva de atún rojo Thunnus thynnus (Linnaeus, 1758). Bol. Inst. Español. Oceangr. 2002, 18, 99–107. [Google Scholar]
- Natalia, Y.; Hashim, R.; Ali, A.; Chong, A. Characterization of digestive enzymes in a carnivorous ornamental fish, the Asian bony tongue Scleropages formosus (Osteoglossidae). Aquaculture 2004, 233, 305–320. [Google Scholar] [CrossRef]
- Sáenz de Rodrigáñez, M.; Alarcón, F.J.; Martínez, M.I.; Ruiz, F.; Díaz, M.; Moyano, F.J. Caracterización de las proteasas digestivas del lenguado senegalés Solea senegalensis Kaup, 1858. Bol. Inst. Español. Oceangr. 2005, 21, 95–104. [Google Scholar]
- Bezerra, S.R.; Lins, J.F.; Alencar, B.R.; Paiva, M.G.; Chaves, E.C.; Coelho, C.B.; Carvalho, B.L. Alkaline proteinase from intestine of Nile tilapia (Oreochromis niloticus). Process Biochem. 2005, 40, 1829–1834. [Google Scholar] [CrossRef]
- Wang, H.; Wang, Y.; Wang, Q.; Xue, C.; Sun, M. Purification and characterization of stomach protease from the turbot (Scophthalmus maximus L.). Fish Physiol. Biochem. 2006, 32, 179–188. [Google Scholar] [CrossRef]
- Perales-García, N. Ontogenia enzimática de la mojarra tenguayaca Petenia splendida. Master’s Thesis, Universidad Juárez Autónoma de Tabasco, Villahermosa, Tabasco, México, 24 November 2010; p. 94. [Google Scholar]
- Nalinanon, S.; Benjakul, S.; Kishimura, H. Biochemical properties of pepsinogen and pepsin from the stomach of albacore tuna (Thunnus alalunga). Food Chem. 2010, 121, 49–55. [Google Scholar] [CrossRef]
- Guerrero-Zarate, R.; Álvarez-González, C.A.; Olvera-Novoa, M.A.; Perales-García, N.; Frías-Quintana, C.A.; Martínez-García, R.; Contreras-Sánchez, W.M. Partial characterization of digestive proteases in tropical gar Atractosteus tropicus juveniles. Fish Physiol. Biochem. 2014, 40, 1021–1029. [Google Scholar] [CrossRef] [PubMed]
- Anson, M.L. The estimation of pepsin, tripsin, papain and cathepsin with hemoglobin. J. Gen. Physiol. 1938, 22, 79–89. [Google Scholar] [CrossRef] [PubMed]
- García-Carreño, L.F.; Albuquerque-Cavalcanti, C.; Navarrete del Toro, A.M.; Zaniboni-Filho, E. Digestive proteinases of Brycon orbignyanus (Characidae, Teleostei): Characteristics and effects or protein quality. Comp. Biochem. Physiol. 2002, 132, 343–352. [Google Scholar] [CrossRef]
- Yúfera, M.; Darias, M.J. Changes in the gastrointestinal pH from larvae to adult in Senegal sole (Solea senegalensis). Aquaculture 2007, 267, 94–99. [Google Scholar] [CrossRef]
- Chong, A.S.C.; Hashim, R.; Chow-Yang, L.; Ali, A.B. Partial characterization and activities of proteases from the digestive tract of discus fish (Symphysodon aequifasciata). Aquaculture 2002, 203, 321–333. [Google Scholar] [CrossRef]
- Tramati, C.; Savona, B.; Mazzola, A. A study of the pattern of digestive enzymes in Diplodus puntazzo (Cetti, 1777) (Osteichthyes, Sparidae): Evidence for the definition of nutritional protocols. Aquac. Int. 2005, 13, 89–95. [Google Scholar] [CrossRef]
- Tanji, M.; Yakabe, E.; Kageyama, T.; Shin-ichi, Y.; Ichinose, M.; Miki, K.; Ito, H.; Takahashi, K. Purification and characterization of pepsinogens from the gastric mucosa of African coelacanth, Latimeria chalumnae, and properties of the major pepsins. Comp. Biochem. Physiol. 2007, 146, 412–420. [Google Scholar] [CrossRef] [PubMed]
- Nalinanon, S.; Benjakul, S.; Visessanguan, W.; Kishimura, H. Tuna Pepsin: Characteristics and its use for collagen extraction from the skin of threadfin bream (Nemipterus spp.). J. Food Sci. 2008, 73, C413–C419. [Google Scholar] [CrossRef] [PubMed]
- Furné, M.; Hidalgo, M.C.; López, A.; García-Gallego, M.; Morales, A.E.; Domezain, A.; Domezainé, J.; Sanz, A. Digestive enzyme activities in Adriatic sturgeon Acipenser naccarii and rainbow trout Oncorhynchus mykiss. A comparative study. Aquaculture 2005, 250, 391–398. [Google Scholar] [CrossRef]
- Álvarez-González, C.A. Actividad enzimática digestiva y evaluación de dietas para el destete de larvas de la cabrilla arenera Paralabrax maculatofasciatus (Percoidei: serranidae). Ph.D. Thesis, Centro Interdisciplinario de Ciencias Marinas, La Paz, Baja California Sur, México, 28 November 2003. [Google Scholar]
- Kumar, S.; García-Carreño, F.L.; Chakrabarti, R.; Toro, M.A.; Córdova-Murueta, J.H. Digestive proteases of three carps Catla catla, Labeo rohita and Hypophthalmichthys molitrix: Partial characterization and protein hydrolysis efficiency. Aquac. Nutr. 2007, 13, 381–388. [Google Scholar] [CrossRef]
- Matus, P.A.; Rosas, A.; Lazo, J.P.; Viana, M.T. Partial characterization of the digestive enzymes of Pacific bluefin tuna Thunnus orientalis under culture conditions. Fish Physiol. Biochem. 2007, 33, 223–231. [Google Scholar]
- Liu, Z.Y.; Wang, Z.; Xu, S.Y.; Xu, L.N. Partial characterization and activity distribution of proteases along the intestine of grass carp, Ctenopharyngodon idella (Val.). Aquac. Nutr. 2008, 14, 31–39. [Google Scholar] [CrossRef]
- Li, J.S.; Li, J.L.; Wu, T. Ontogeny of protease, amylase and lipase in the alimentary tract of hybrid juvenile tilapia (Oreochromis niloticus × Oreochromis aureus). Fish Physiol. Biochem. 2006, 32, 295–303. [Google Scholar]
- Alarcón, F.J.; Díaz, M.; Moyano, F.J.; Abellan, E. Characterization and functional properties of digestive proteases in two sparids; gilthead seabream (Sparus aurata) and common dentex (Dentex dentex). Fish Physiol. Biochem. 1998, 19, 257–267. [Google Scholar] [CrossRef]
- Nalinanon, S.; Benjakul, S.; Kishimura, H. Purifcation and biochemical properties of pepsins from the stomach of skipjack tuna (Katsuwonus pelamis). Eur. Food Res. Technol. 2010, 231, 259–269. [Google Scholar] [CrossRef]
- Alencar, B.R.; Biondi, M.M.; Paiva, M.V.; Carvalho-Junior, B.L.; Bezerra, S.R. Alkaline proteases from the digestive tract of four tropical fishes. Braz. J. Food Technol. 2003, 6, 279–284. [Google Scholar]
- Uscanga, A.; Moyano, F.J.; Álvarez, C.A. Assessment of enzymatic efficiency on protein digestion in the tilapia Oreochromis niloticus. Fish Physiol. Biochem. 2010, 36, 1079–1085. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Liu, G.; Huang, Y.; Weng, L.; Hara, K.; Su, W.; Cao, M. Pepsinogens and pepsins from mandarin fish (Siniperca chuatsi). J. Agric. Food Chem. 2008, 56, 5401–5406. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Yáñez, F.J.; Pacheco-Aguilar, R.; García-Carreño, F.L.; Navarrete-Del Toro, M.A. Isolation and characterization of trypsin from pyloric caeca of Monterey sardine Sagax caerulea caerulea. Comp. Biochem. Physiol. B 2005, 140, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Ben, K.H.; Jellouli, K.; Souissi, N.; Ghorbel, S.; Barkia, A.; Nasri, M. Purification and characterization of three trypsin isoforms from viscera of sardinelle (Sardinella aurita). Fish Physiol. Biochem. 2011, 37, 123–133. [Google Scholar]
- Castillo-Yáñez, F.J.; Pacheco-Aguilar, R.; Lugo-Sánchez, M.E.; García-Sánchez, G.; Quintero-Reyes, I.E. Biochemical characterization of an isoform of chymotrypsin from the viscera of Monterey sardine (Sardinops sagax caerulea), and comparison with bovine chymotrypsin. Food Chem. 2009, 112, 634–639. [Google Scholar] [CrossRef]
- Simpson, B.K. Digestive Proteases from Marine Animals. In Seafood Enzymes; Haard, N.F., Simpson, B.K., Eds.; Marcel Dekker: New York, NY, USA, 2000; pp. 191–213. [Google Scholar]
- Dunn, B.M. Determination of protease mechanism. In Proteolytic Enzymes: A Practical Approach; Beynon, R.J., Bond, J.S., Eds.; IRL Press: Oxford, UK, 1989; pp. 57–58. [Google Scholar]
- Walter, H.E. Proteinases: Methods with hemoglobin, casein and azocoll as substrates. In Methods of Enzymatic Analysis; Bergmeyer, H.J., Ed.; Verlag Chemie: Weinheim, Germany, 1984; pp. 270–277. [Google Scholar]
- Kunitz, M. Crystalline soybean trypsin inhibitor II. General properties. J. Gen. Physiol. 1947, 30, 291–310. [Google Scholar] [CrossRef] [PubMed]
- Stauffer, C. Enzyme Assays for Food Scientists; Van Nostand Reinhold/AVI: New York, NY, USA, 1989; p. 317. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Cárdenas, L.; Álvarez-González, C.A.; Hernández-Almeida, O.U.; Frías-Quintana, C.A.; Ponce-Palafox, J.T.; Castillo-Vargasmachuca, S. Partial Characterization of Digestive Proteases in the Green Cichlid, Cichlasoma beani. Fishes 2017, 2, 4. https://doi.org/10.3390/fishes2010004
Martínez-Cárdenas L, Álvarez-González CA, Hernández-Almeida OU, Frías-Quintana CA, Ponce-Palafox JT, Castillo-Vargasmachuca S. Partial Characterization of Digestive Proteases in the Green Cichlid, Cichlasoma beani. Fishes. 2017; 2(1):4. https://doi.org/10.3390/fishes2010004
Chicago/Turabian StyleMartínez-Cárdenas, Leonardo, Carlos A. Álvarez-González, Oscar U. Hernández-Almeida, Carlos A. Frías-Quintana, Jesús T. Ponce-Palafox, and Sergio Castillo-Vargasmachuca. 2017. "Partial Characterization of Digestive Proteases in the Green Cichlid, Cichlasoma beani" Fishes 2, no. 1: 4. https://doi.org/10.3390/fishes2010004






