A Role for Soluble IL-6 Receptor in Osteoarthritis
Abstract
:1. Introduction
2. Cartilage Alterations in Osteoarthritis (OA)
3. “Classical” IL-6 Signaling Versus IL-6 Trans-Signaling
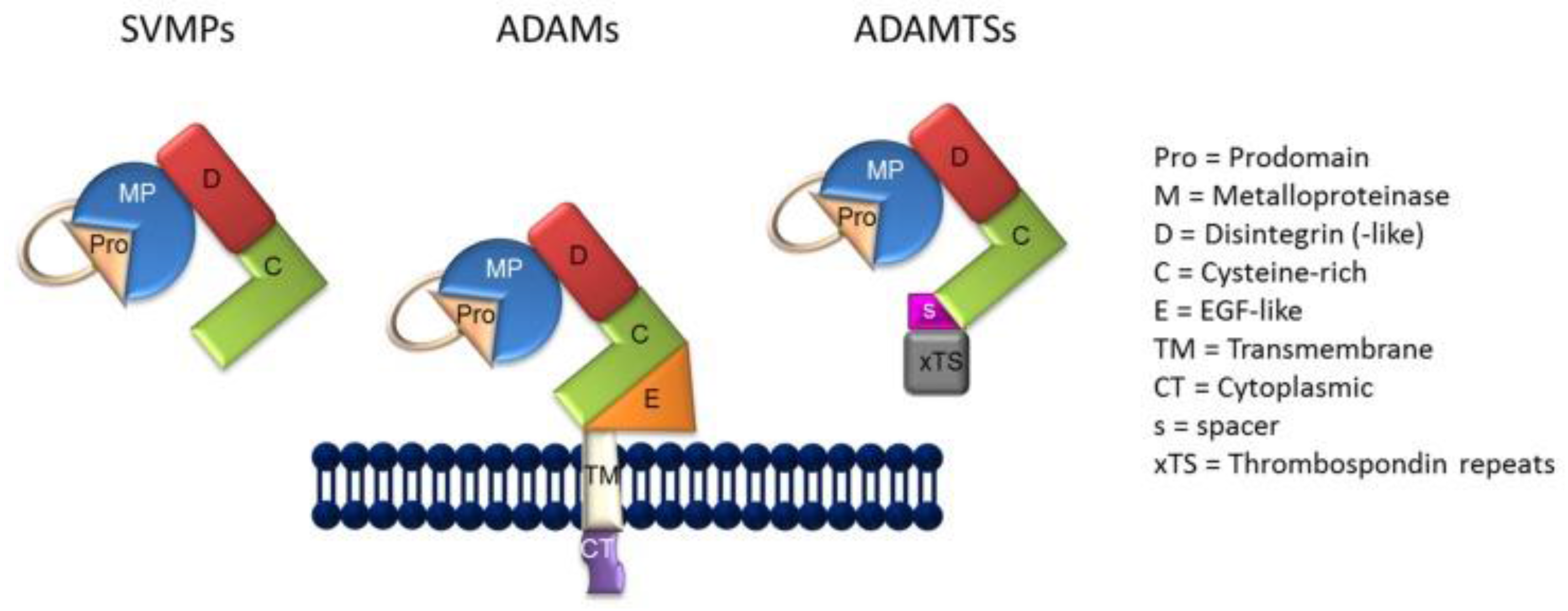
4. Metzincin Proteases Contribute to the Formation sIL-6R
5. A Role for the ADAMs in OA?
6. Conclusions and Future Perspective
Acknowledgments
Conflicts of Interest
References
- Denko, C.W.; Malemud, C.J. Metabolic disturbances and synovial joint responses in osteoarthritis. Front. Biosci. 1999, 4, d686–d693. [Google Scholar] [CrossRef] [PubMed]
- Cicuttini, F.M.; Wluka, A.E. Osteoarthritis: Is OA a mechanical or systemic disease? Nat. Rev. Rheumatol. 2014, 10, 515–516. [Google Scholar] [PubMed]
- Sellam, J.; Berenbaum, F. The role of synovitis in pathophysiology and clinical symptoms of osteoarthritis. Nat. Rev. Rheumatol. 2010, 6, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Hügle, T.; Geurts, J.; Nüesch, G.; Müller-Gerbi, M.; Valderrabono, V. Aging and osteoarthritis: An inevitable encounter? J. Aging Res. 2012, 2012, 950192. [Google Scholar] [PubMed]
- Meszaros, E.; Malemud, C.J. Prospects for treating osteoarthritis: Enzyme-protein interactions regulating matrix metalloproteinase activity. Ther. Adv. Chronic Dis. 2012, 3, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, J.-P.; Martel-Pelletier, J.; Abramson, S.B. Osteoarthritis, an inflammatory disease: Potential implications for the selection of new therapeutic targets. Arthritis Rheum. 2001, 44, 1237–1247. [Google Scholar] [PubMed]
- Malemud, C.J.; Islam, N.; Haqqi, T.M. Pathophysiological mechanisms in osteoarthritis lead to novel therapeutic strategies. Cells Tissues Organs 2003, 174, 34–48. [Google Scholar] [PubMed]
- Goldring, M.B.; Otero, M. Inflammation in osteoarthritis. Curr. Opin. Rheumatol. 2011, 23, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Malemud, C.J. The biological basis of osteoarthritis: State of the evidence. Curr. Opin. Rheumatol. 2015, 27, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Malemud, C.J.; Schulte, M.E. Is there a final common pathway for arthritis? Future Rheumatol. 2008, 3, 253–268. [Google Scholar]
- Scanzello, C.R.; Goldring, S.R. The role of synovitis in osteoarthritis pathogenesis. Bone 2012, 51, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Berenbaum, F. Osteoarthritis as an inflammatory disease (osteoarthritis is not osteoarthrosis!). Osteoarthr. Cartil. 2013, 21, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Sakkas, L.I.; Platsoucas, P.D. The role of T cells in the pathogenesis of osteoarthritis. Arthritis Rheum. 2007, 56, 409–424. [Google Scholar] [CrossRef] [PubMed]
- De Lange-Brokkar, B.J.; Ioan-Facsinay, A.; van Osch, G.J.; Zuurmond, A.M.; Schoones, J.; Toes, R.E.; Huizinga, T.W.; Kloppenberg, M. Synovial inflammation, immune cells, and their cytokines in osteoarthritis: A review. Osteoarthr. Cartil. 2012, 20, 1484–1499. [Google Scholar] [CrossRef] [PubMed]
- Haseeb, A.; Haqqi, T.M. Immunopathogenesis of osteoarthritis. Clin. Immunol. 2013, 146, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, M.; Martel-Pelletier, J.; Lajeunesse, D.; Pelletier, J.-P.; Fahmi, H. Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat. Rev. Rheumatol. 2011, 7, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Gargiulo, S.; Gamba, P.; Poli, G.; Leonarduzzi, G. Metalloproteinases and metalloproteinase inhibitors in age-related diseases. Curr. Pharm. Des. 2014, 20, 2993–3018. [Google Scholar] [CrossRef] [PubMed]
- Malemud, C.J. Matrix Metalloproteinases and Tissue Remodeling in Health and Disease Part II. Target Tissues and Therapy; Elsevier: Amsterdam, The Netherlands, 2017; pp. 306–325. [Google Scholar]
- Verma, P.; Dakal, K. ADAMTS-4 and ADAMTS-5: Key enzymes in osteoarthritis. J. Cell. Biochem. 2011, 112, 3507–3514. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.Y.; Chanalaris, A.; Troeberg, L. ADAMTS and ADAM metalloproteinases in osteoarthritis—Looking beyond the ‘usual suspects’. Osteoarthr. Cartil. 2017, 25, 1000–1009. [Google Scholar] [CrossRef] [PubMed]
- Huard, X.; Goldring, M.B.; Berenbaum, F. Homeostatic mechanisms in articular cartilage and role of inflammation in osteoarthritis. Curr. Rheumatol. Rep. 2013, 15, 375. [Google Scholar] [CrossRef] [PubMed]
- Wylie, M.A.; Malemud, C.J. Perspective: Deregulation of apoptosis in arthritis by altered signal transduction. Int. J. Clin. Rheumatol. 2013, 8, 483–490. [Google Scholar] [CrossRef]
- Malemud, C.J. Osteoarthritis, Inflammation and Degradation: A Continuum; Volume, 70: Biomedical and Health Research; IOS Press: Amsterdam, The Netherlands, 2007; pp. 99–117. [Google Scholar]
- Malemud, C.J.; Pearlman, E. Targeting JAK/STAT signaling pathway in inflammatory diseases. Curr. Signal Transduct. Ther. 2009, 4, 201–221. [Google Scholar] [CrossRef]
- Malemud, C.J. Drug Discovery; InTech Publishing: Rijeka, Croatia, 2013; pp. 373–411. [Google Scholar]
- Malemud, C.J. Negative regulators of JAK/STAT signaling in rheumatoid arthritis and osteoarthritis. Int. J. Mol. Sci. 2017, 18, 484. [Google Scholar] [CrossRef] [PubMed]
- Melas, I.N.; Chairakaki, A.D.; Chatzopoulou, E.I.; Messinis, D.E.; Katopodi, T.; Pilaka, V.; Samara, S.; Mitsos, A.; Dailana, Z.; Kollia, P.; et al. Modeling of signaling pathways in chondrocytes based on phosphoproteomics and cytokine release data. Osteoarthr. Cartil. 2014, 22, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Huang, X.; Li, L.; Huang, H.; Xu, R.; Luyten, W. Insights on biology and pathology of HIF-1α/-2α, TGFβ/BMP, Wnt/β-catenin and NF-κB pathways in osteoarthritis. Curr. Pharm. Des. 2012, 18, 3293–3312. [Google Scholar] [CrossRef] [PubMed]
- Rigoglou, S.; Papavassilou, A.G. The NF-κB signaling pathway in osteoarthritis. Int. J. Biochem. Cell. Biol. 2013, 45, 2580–2584. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, A.I.; Beekhuizen, M.T.; Ct Hart, M.C.; Radstake, T.R.; Dhert, W.A.; Saris, W.B.; van Osch, G.J.; Creemers, L.B. Cytokine profiles in the joint depend on pathology, but are different between synovial fluid, cartilage tissue and cultured chondrocytes. Arthritis Res. Ther. 2014, 16, 441. [Google Scholar] [CrossRef] [PubMed]
- DeBoer, T.N.; van Spil, W.E.; Huisman, A.M.; Polak, A.A.; Bijlsma, J.W.; Lafeber, F.P.; Mastbergen, F.C. Serum adipokines in osteoarthritis: Comparison with controls and relationship with local parameters of synovial inflammation and cartilage damage. Osteoarthr. Cartil. 2012, 20, 846–853. [Google Scholar] [CrossRef] [PubMed]
- Livshits, G.; Zhai, G.; Hart, D.J.; Kato, B.S.; Wang, H.; Williams, F.M.; Spector, T.D. Interleukin-6 is a significant predictor of radiographic knee osteoarthritis: The Chingford study. Arthritis Rheumatol. 2009, 60, 2037–2045. [Google Scholar] [CrossRef] [PubMed]
- Blumenfeld, O.; Williams, F.M.; Valdes, A.; Hart, D.J.; Malkin, I.; Spector, T.D.; Livshits, G. Association of interleukin-6 polymorphisms with hand osteoarthritis and hand osteoporosis. Cytokine 2014, 69, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Iannone, F.; Lapadula, G. The pathophysiology of osteoarthritis. Aging Clin. Exp. Res. 2003, 15, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Goldring, M.; Goldring, S. Osteoarthritis. J. Cell. Physiol. 2007, 213, 626–634. [Google Scholar] [CrossRef] [PubMed]
- Sandell, L.; Aigner, T. Articular cartilage and changes in arthritis. An introduction: Cell biology of osteoarthritis. Arthritis Res. 2001, 3, 107–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hulejová, H.; Baresová, V.; Klèzl, Z.; Polanská, M.; Adam, M.; Senolt, L. Increased level of cytokines and matrix metalloproteinases in osteoarthritic subchondral bone. Cytokine 2007, 38, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Hollander, A.P.; Pidoux, I.; Reiner, A.; Rorabeck, C.; Bourne, R.; Poole, A.R. Damage to type II collagen in aging and osteoarthritis starts at the articular surface, originates around chondrocytes, and extends into the cartilage with progressive degeneration. J. Clin. Investig. 1995, 96, 2859–2869. [Google Scholar] [CrossRef] [PubMed]
- Burrage, P.S.; Mix, K.S.; Brinckerhoff, C.E. Matrix metalloproteinases: Role in arthritis. Front. Biosci. 2006, 11, 529–543. [Google Scholar] [CrossRef] [PubMed]
- Burrage, P.S.; Brinckerhoff, C.E. Molecular targets in osteoarthritis: Metalloproteinases and their inhibitors. Curr. Drug Targets 2007, 8, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Struglics, A.; Larsson, S.; Pratta, M.A.; Kumar, S.; Lark, M.W.; Lohmander, L.S. Human osteoarthritis synovial fluid and joint cartilage contain both aggrecanase- and matrix metalloproteinase-generated aggrecan fragments. Osteoarthr. Cartil. 2006, 14, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Durigova, M.; Roughley, P.J.; Mort, J.S. Mechanism of proteoglycan aggregate degradation in cartilage stimulated with oncostatin M. Osteoarthr. Cartil. 2008, 16, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Yuan, X.M.; Yao, Q.; Peng, L.B. OSM is overexpressed in knee osteoarthritis and Notch signaling is involved in the effects of OSM on MC3T3-E1 cell proliferation and differentiation. Int. J. Mol. Med. 2015, 35, 1755–1760. [Google Scholar] [CrossRef] [PubMed]
- Greene, M.A.; Loesser, R.F. Function of the chondrocyte PI-3 kinase-Akt signaling pathway is stimulus dependent. Osteoarthr. Cartil. 2015, 23, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Mueller, M.B.; Tuan, R.S. Anabolic/catabolic balance in the pathogenesis of osteoarthritis: Identifying molecular targets. PMR 2011, 3, S3–S11. [Google Scholar] [CrossRef] [PubMed]
- Hedbom, E.; Häuselmann, H.J. Molecular aspects of pathogenesis in osteoarthritis: The role of inflammation. Cell. Mol. Life Sci. 2002, 59, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Haan, C.; Is’harc, H.; Hermanns, H.M.; Schmitz-Van de Leur, H.; Kerr, I.M.; Heinrich, P.C.; Grötzinger, J.; Behrmann, I. Mapping of a region within the N terminus of Jak1 involved in cytokine receptor interaction. J. Biol. Chem. 2001, 276, 37451–37458. [Google Scholar] [CrossRef] [PubMed]
- Haan, C.; Hermanns, H.M.; Heinrich, P.C.; Behrmann, I. A single amino acid substitution (Trp666→Ala) in the interbox1/2 region of the interleukin-6 signal transducer gp130 abrogates binding of JAK1, and dominantly impairs signal transduction. Biochem. J. 2000, 349, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Haan, C.; Heinrich, P.C.; Behrmann, I. Structural requirements of the interleukin-6 signal transducer gp130 for its interaction with Janus kinase 1: The receptor is crucial for kinase activation. Biochem. J. 2002, 361, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Hallek, M.; Neumann, C.; Schaffer, M.; Danhauser-Riedl, S.; von Bubnoff, N.; de Vos, G.; Druker, B.J.; Yasukawa, K.; Griffin, J.D.; Emmerich, B. Signal transduction of interleukin-6 involves tyrosine phosphorylation of multiple cytosolic proteins and activation of Src-family kinases Fyn, Hck, and Lyn in multiple myeloma cell lines. Exp. Hematol. 1997, 25, 1367–1377. [Google Scholar] [PubMed]
- Schaeffer, M.; Schneiderbauer, M.; Weidler, S.; Tavares, R.; Warmuth, M.; de Vos, G.; Hallek, M. Signaling through a novel domain of gp130 mediates cell proliferation and activation of Hck and Erk kinases. Mol. Cell. Biol. 2001, 21, 8068–8081. [Google Scholar] [CrossRef] [PubMed]
- Malemud, C.J. PI3K/Akt/PTEN/mTOR signaling: A fruitful target for inducing cell death in rheumatoid arthritis? Future Med. Chem. 2015, 7, 1137–1147. [Google Scholar] [CrossRef] [PubMed]
- Oberg, H.-H.; Wesch, D.; Grüssel, S.; Rose-John, S.; Kabelitz, D. Differential expression of CD126 and CD130 mediates different STAT-3 phosphorylation in CD4+CD25− and CD25high regulatory T cells. Int. Immunol. 2006, 18, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Gauldie, J.; Richards, C.; Harnish, D.; Lansdorp, P.; Baumann, H. Interferon β2/B-cell stimulatory factor type 2 shares identity with monocyte-derived hepatocyte-stimulating factor and regulates the major acute phase protein response in liver cells. Proc. Natl. Acad. Sci. USA 1987, 84, 7251–7255. [Google Scholar] [CrossRef]
- Schuster, B.; Kovaleva, M.; Sun, Y.; Regenhard, P.; Matthews, V.; Grötzinger, J.; Rose-John, S.; Kallen, K.J. Signaling of human ciliary neurotrophic factor (CNTF) revisited. The interleukin-6 receptor can serve as an α-receptor for CNTF. J. Biol. Chem. 2003, 278, 9528–9535. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.-S.; Choi, J.H.; Lee, S.Y.; Park, Y.-H.; Park, K.-Y.; Lee, J.Y.; Kim, J.; Gajulapati, V.; Goo, J.-I.; Singh, S.; et al. A novel small-molecule inhibitor targeting the IL-6 receptor β subunit, glycoprotein 130. J. Immunol. 2015, 195, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Marotte, H.; Kwan, K.; Ruth, J.H.; Campbell, P.L.; Rabquer, B.J.; Pakozdi, A.; Koch, A.E. Epigallocatechin-3-gallate inhibits IL-6 synthesis and suppresses transsignaling by enhancing soluble gp130 production. Proc. Natl. Acad. Sci. USA 2008, 105, 14692–14697. [Google Scholar] [CrossRef] [PubMed]
- Mülberg, J.; Schooltink, H.; Stoyan, T.; Günther, M.G.; Lutz, B.; Mackiewicz, B.A.; Heinrich, P.C.; Rose-John, S. The soluble interleukin-6 receptor is generated by shedding. Eur. J. Immunol. 1993, 23, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Lust, J.A.; Donovan, K.A.; Kline, M.P.; Greipp, P.R.; Kyle, R.A.; Maihle, N.J. Isolation of an mRNA encoding a soluble form of the human interleukin-6 receptor. Cytokine 1992, 4, 96–100. [Google Scholar] [CrossRef]
- Taga, T.; Hibi, M.; Hirata, Y.; Yamasaki, K.; Yasukawa, K.; Matsuda, T.; Hirano, T.; Kishimoto, T. Interleukin-6 triggers the association of its receptor with a possible signal transducer, gp130. Cell 1989, 58, 573–581. [Google Scholar] [CrossRef]
- Aida, Y.; Honda, K.; Tanigawa, S.; Nakayama, G.; Matsumura, H.; Suzuki, N.; Shimizu, O.; Takeichi, O.; Makimura, M.; Maeno, M. IL-6 and soluble IL-6 receptor stimulate the production of MMPs and their inhibitors via JAK-STAT and ERK-MAPK signalling in human chondrocytes. Cell. Biol. Int. 2012, 36, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Scheller, J.; Garbers, C.; Rose-John, S. Interleukin-6: From basic biology to selective blockade of pro-inflammatory activities. Semin. Immunol. 2014, 26, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Heink, S.; Yogev, N.; Garbers, C.; Herwerth, M.; Aly, L.; Gasperi, C.; Husterer, V.; Croxford, A.L.; Möller-Hackbarth, K.; et al. Trans-presentation of interleukin-6 by dendritic cells is required for priming pathogenic TH17 cells. Nat. Immunol. 2017, 18, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Garbers, C.; Aparicio-Siegmund, S.; Rose-John, S. The IL-6/gp130/STAT3 signaling axis: Recent advances towards specific inhibition. Curr. Opin. Immunol. 2015, 34, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Nowell, M.A.; Richards, P.J.; Horiuchi, S.; Yamamoto, N.; Rose-John, S.; Topley, N.; Williams, A.S.; Jones, S.A. Soluble IL-6 receptor governs IL-6 activity in experimental arthritis: Blockade of arthritis severity by soluble glycoprotein 130. J. Immunol. 2003, 171, 3202–3209. [Google Scholar] [CrossRef] [PubMed]
- Campbell, I.L.; Erta, M.; Lim, S.L.; Frausto, R.; May, U.; Rose-John, S.; Scheller, J.; Hidalgo, J. Trans-signaling is a dominant mechanism for the pathogenic actions of interleukin-6 in the brain. J. Neurosci. 2014, 34, 2503–2513. [Google Scholar] [CrossRef] [PubMed]
- Hirahara, K.; Atsushi, O.; Villarino, A.V.; Bonelli, M.; Sciumè, G.; Laurence, A.; Sun, H.-W.; Brooks, S.R.; Vahedi, G.; Shih, H.-Y.; et al. Asymmetric action of STAT transcription factors drives transcriptional outputs and cytokine specificity. Immunity 2017, 42, 877–889. [Google Scholar] [CrossRef] [PubMed]
- Rose-John, S.; Waetzig, G.H.; Scheller, J.; Grötzinger, J.; Seegert, D. The IL-6/sIL-6R complex as a novel target for therapeutic approaches. Expert Opin. Ther. Targets 2007, 11, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Klein, T.; Bischoff, R. Active metalloproteases of the A Disintegrin and Metalloprotease (ADAM) family; biological function and structure. J. Proteome Res. 2011, 10, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Rose-John, S. The soluble interleukin-6 receptor and related proteins. Best Pract. Res. Clin. Endocrinol. Metab. 2015, 29, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Bridges, L.C.; Bowditch, R.D. ADAM-integrin interactions: Potential integrin regulated ectodomain shedding activity. Curr. Pharm. Des. 2005, 11, 837–847. [Google Scholar] [CrossRef] [PubMed]
- Saftig, P.; Reiss, K. The “A Disintegrin and Metalloproteases” ADAM10 and ADAM17: Novel drug targets with therapeutic potential? Eur. J. Cell. Biol. 2011, 90, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, M.; Herrlich, A.; Herrlich, P. Who decides when to cleave an ectodomain? Trends Biochem. Sci. 2013, 38, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Giebeler, N.; Zigrino, P. A disintegrin and metalloprotease (ADAM): Historical overview of their functions. Toxins 2016, 8, 122. [Google Scholar] [CrossRef] [PubMed]
- Chalaris, A.; Garbers, C.; Rabe, B.; Rose-John, S.; Scheller, J. The soluble interleukin 6 receptor: Generation and role in inflammation and cancer. Eur. J. Cell. Biol. 2011, 90, 484–494. [Google Scholar] [CrossRef] [PubMed]
- Rose-John, S. ADAM17, shedding, TACE as therapeutic targets. Pharmacol. Res. 2013, 71, 19–22. [Google Scholar] [CrossRef] [PubMed]
- Endres, K.; Fahrenholtz, F. Regulation of α-secretase ADAM10 expression and activity. Exp. Brain Res. 2012, 217, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, N.; Meyer, D.; Mauermann, A.; von der Heyde, J.; Wolf, J.; Schwarz, J.; Knittler, K.; Murphy, G.; Michalek, M.; Garbers, C.; et al. Shedding of endogenous interleukin-6 receptor (IL-6R) is governed by a distintegrin and metalloprotease (ADAM) proteases while a full-length IL-6R isoform localizes to circulating microvesicles. J. Biol. Chem. 2015, 290, 26059–26071. [Google Scholar] [CrossRef] [PubMed]
- Klein, T.; Bischoff, R. Physiology and pathophysiology of matrix metalloproteases. Amino Acids 2011, 41, 271–290. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Mukerjee, S.; Silva-Alves, C.R.; Carvalho-Galvặo, A.; Cruz, J.C.; Balarini, C.M.; Braga, V.A.; Lazartiques, E.; Franҫa-Silva, M.S. A disintegrin and metalloprotease 17 in the cardiovascular and central nervous systems. Front. Physiol. 2016, 7, 469. [Google Scholar] [CrossRef] [PubMed]
- Scheller, J.; Chalaris, A.; Garbers, C.; Rose-John, S. ADAM17: A molecular switch to control inflammation and tissue regeneration. Trends Immunol. 2011, 32, 880–887. [Google Scholar] [CrossRef] [PubMed]
- Lisi, S.; D’Amore, M.; Sisto, M. ADAM17 at the interface between inflammation and autoimmunity. Immunol. Lett. 2014, 162, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Chalaris, A.; Gewiese, J.; Paliga, K.; Fleig, L.; Schneede, A.; Krieger, K.; Rose-John, S.; Scheller, J. ADAM17-mediated shedding of the IL-6R induces cleavage of the membrane stub by γ-secretase. Biochim. Biophys. Acta 2010, 1803, 234–245. [Google Scholar] [CrossRef] [PubMed]
- Crawford, H.C.; Dempsey, P.J.; Brown, G.; Adam, L.; Moss, M.L. ADAM10 as a therapeutic target for cancer and inflammation. Curr. Pharm. Des. 2009, 15, 2288–2299. [Google Scholar] [CrossRef] [PubMed]
- Gibb, D.R.; Saleem, S.J.; Chaimowitz, N.S.; Mathews, J.; Conrad, D.H. The emergence of ADAM10 as a regulator of lymphocyte development and autoimmunity. Mol. Immunol. 2011, 48, 1319–1327. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, A.; Hundhausen, C.; Lambert, M.H.; Broadway, N.; Andrews, R.C.; Bickett, D.M.; Lessnitzer, M.A.; Becherer, J.D. Metalloprotease inhibitors for the disintegrin and metalloproteases ADAM10 and ADAM17 that differentially block constitutive and phorbol ester-inducible shedding from cell surface molecules. Comb. Chem. High Throughput Screen. 2005, 8, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Jangouk, P.; Dahmel, T.; Meyer Zu Hörst, G.; Ludwig, A.; Lehmann, H.C.; Kieseier, B.C. Involvement of ADAM10 in axonal outgrowth and myelination of the peripheral nerve. Glia 2009, 57, 1765–1774. [Google Scholar] [CrossRef] [PubMed]
- Durairajan, S.S.; Liu, L.F.; Lu, J.H.; Koo, I.; Maruyama, K.; Chung, S.K.; Huang, J.D.; Li, M. Stimulation of non-amyloidogenic processing of amyloid-ß protein precursor by cryptotanshinone involves activation and translocation of ADAM10 and PKC-α. J. Alzheimers Dis. 2011, 25, 245–262. [Google Scholar] [PubMed]
- Dreymueller, D.; Martin, C.; Kogel, T.; Pruessmeyer, J.; Hess, F.M.; Horiuchi, K.; Uhlig, S.; Ludwig, A. Lung epithelial ADAM17 regulates the acute inflammatory response to lipopolysaccharide. EMBO Mol. Med. 2012, 4, 412–423. [Google Scholar] [CrossRef] [PubMed]
- Maretzky, T.; Swendeman, S.; Mogollon, E.; Weskamp, G.; Sahin, U.; Reiss, K.; Blobel, C.P. Characterization of the catalytic properties of the membrane-anchored metalloprotease ADAM9 in cell-based assays. Biochem. J. 2017, 474, 1467–1479. [Google Scholar] [CrossRef] [PubMed]
- Li, N.G.; Shi, Z.H.; Tang, Y.P.; Wei-Li; Lian-Yin; Duan, J.A. Discovery of selective small molecule TACE inhibitors for the treatment of rheumatoid arthritis. Curr. Med. Chem. 2012, 19, 2924–2956. [Google Scholar] [PubMed]
- Campard, D.; Vasse, M.; Rose-John, S.; Poyer, F.; Lamacz, M.; Vannier, J.P. Multilevel regulation of IL-6R by IL-6-sIL-6R fusion protein according to the primitiveness of peripheral blood-derived CD133+ cells. Stem Cells 2006, 24, 1302–1314. [Google Scholar] [CrossRef] [PubMed]
- Garbers, C.; Monhasery, N.; Aparicio-Siegmund, S.; Lokau, J.; Baran, P.; Nowell, M.A.; Jones, S.A.; Rose-John, S.; Scheller, J. The interleukin-6 receptor, Asp358Ala single nucleotide polymorphism rs2228145 confers increased proteolytic conversion rates by ADAM proteases. Biochim. Biophys. Acta 2014, 1842, 1485–1494. [Google Scholar] [CrossRef] [PubMed]
- Muoh, O.; Malemud, C.J.; Askari, A.D. Clinical significance and implications of genetic and genomic studies in patients with osteoarthritis. Adv. Genom. Genet. 2014, 4, 193–206. [Google Scholar]
- Oldefest, M.; Düsterhöft, S.; Desel, C.; Thysen, S.; Fink, C.; Rabe, B.; Lories, R.; Grötzinger, J.; Lorenzen, I. Secreted Frizzled-related protein 3 (sFRP3)-mediated suppression of interleukin-6 receptor release by A disintegrin and metalloprotease 17 (ADAM17) is abrogated in the osteoarthritis-associated rare double variant of sFRP3. Biochem. J. 2015, 468, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Yan, I.; Schwarz, J.; Lücke, K.; Schumacher, N.; Schumacher, V.; Schmidt, S.; Rabe, B.; Saftig, P.; Donners, M.; Rose-John, S.; et al. ADAM17 controls IL-6 signaling by cleavage of the murine IL-6Rα from the cell surface of leukocytes during inflammatory responses. J. Leukoc. Biol. 2016, 99, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Reithmueller, S.; Somasundaram, P.; Ehlers, J.C.; Hung, C.W.; Flynn, C.M.; Lokau, J.; Agthe, M.; Düsterhöft, S.; Zhu, Y.; Grötzinger, J.; et al. Proteolytic origin of the soluble human IL-6R in vivo and a decisive role of N-glycosylation. PLoS ONE 2017, 15, e2000080. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.; Shukla, M.; Malemud, C.J.; Cowland, J.; Haqqi, T.M. Neutrophil gelatinase-associated lipocalin (NGAL) is produced by IL-1β-stimulated human osteoarthritis chondrocytes and protects MMP-9 from degradation. Osteoarthr. Cartil. 2006, 14, S87–S88. [Google Scholar] [CrossRef]
- Gupta, K.; Shukla, M.; Cowland, J.B.; Malemud, C.J.; Haqqi, T.M. Neutrophil gelatinase-associated lipocalin (NGAL) is expressed in osteoarthritis and forms a complex with matrix metalloproteinase-9. Arthritis Rheum. 2007, 56, 3326–3335. [Google Scholar] [CrossRef] [PubMed]
- Meszaros, E.C.; Dahoud, W.; Mesiano, S.; Malemud, C.J. Blockade of recombinant human IL-6 with tocilizumab suppresses matrix metalloproteinase-9 production in the C28/I2 immortalized human chondrocyte cell line. Integr. Mol. Med. 2015, 2, 304–310. [Google Scholar] [CrossRef] [PubMed]
- Shetty, A.; Hanson, R.; Korsten, P.; Shawagfeh, M.; Arami, S.; Volkov, S.; Vila, O.; Swedla, W.; Shunaigat, A.S.; Smadi, S.; et al. Tocilizumab in the treatment of rheumatoid arthritis and beyond. Drug Des. Dev. Ther. 2014, 8, 349–364. [Google Scholar]
- Malemud, C.J.; Meszaros, E.C.; Wylie, M.A.; Dahoud, W.; Skomorovska-Prokvolit, Y.; Mesiano, S. Matrix metalloproteinase-9 production in immortalized human chondrocyte lines. J. Clin. Cell. Immunol. 2016, 7, 422. [Google Scholar] [CrossRef] [PubMed]
- Kelwick, R.; Desanlis, I.; Wheeler, G.N.; Edwards, D.R. The ADAMTS (a distintegrin and metalloproteinase with thrombospondin motif) family. Genome Biol. 2015, 16, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mimata, Y.; Kamataki, A.; Oikawa, S.; Murakami, K.; Uzuki, M.; Shimamura, T.; Sawai, T. Interleukin-6 upregulates the expression of ADAMTS-4 in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Int. J. Rheumatol. Dis. 2012, 15, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Dreymueller, D.; Ludwig, A. Considerations of inhibition approaches for proinflammatory functions of ADAM proteases. Platelets 2017, 28, 354–361. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akeson, G.; Malemud, C.J. A Role for Soluble IL-6 Receptor in Osteoarthritis. J. Funct. Morphol. Kinesiol. 2017, 2, 27. https://doi.org/10.3390/jfmk2030027
Akeson G, Malemud CJ. A Role for Soluble IL-6 Receptor in Osteoarthritis. Journal of Functional Morphology and Kinesiology. 2017; 2(3):27. https://doi.org/10.3390/jfmk2030027
Chicago/Turabian StyleAkeson, Graham, and Charles J. Malemud. 2017. "A Role for Soluble IL-6 Receptor in Osteoarthritis" Journal of Functional Morphology and Kinesiology 2, no. 3: 27. https://doi.org/10.3390/jfmk2030027



