Five-Year Antimicrobial Susceptibility of Pseudomonas aeruginosa from a Local Tertiary Hospital in Bacolod City, Philippines
Abstract
:1. Introduction
2. Materials and Methods
2.1. Inclusion and Exclusion Criteria
2.2. Isolation and Identification
2.3. Antibiotic Susceptibility Test
2.4. AmpC Screening Test Using Cefoxitin
2.5. Multiple Antibiotic Resistance Index
2.6. Quality Control
2.7. Ethical Consideration
2.8. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hauser, A.R. Pseudomonas. In Antibiotic Basics for Clinicians: Choosing the Right Antibacterial Agent; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007; pp. 129–131. [Google Scholar]
- Dale, D.; Federman, D.; Antman, K.; Atkinson, J.; Cassel, C.; Feldman, M.; Wolinsky, J. ACP Medicine, 2nd ed.; WebMD Inc: New York, NY, USA, 2006. [Google Scholar]
- Aloush, V.; Navon-Venezia, S.; Seiman-Igra, Y.; Cabili, S.; Carmeli, Y. Multi-drug resistant Pseudomonas aeruginosa: risk factors and clinical impact. Antimicrob. Agents Chemother. 2006, 50, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Janner, D. A Clinical Guide to Pediatric Infectious Disease; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2005. [Google Scholar]
- Clinical Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fifth Informational Supplement M100-S25; CLSI: Wayne, PA, USA, 2015. [Google Scholar]
- Roca, I.; Akova, M.; Baquero, F.; Carlet, J.; Cavaleri, M.; Coenen, S.; Cohen, J.; Findlay, D.; Gyssens, I.; Heure, O.E.; et al. The global threat of antimicrobial resistance: Science for intervention. New Microbes New Infect. 2015, 6, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Garba, I.; Lusa, H.; Bawa, E.; Tijjani, M.B.; Aliyu, M.S.; Zango, U.U.; Raji, M.I.O. Antibiotics susceptibility pattern of Pseudomonas aeruginosa isolated from wounds in patients attending Ahmadu Bello University Teaching Hospital, Zaria, Nigeria. Niger. J. Basic Appl. Sci. 2012, 20, 32–34. [Google Scholar]
- Fatima, A.; Naqvi, S.B.; Khaliq, S.A.; Reveen, S.; Jabeen, S. Antimicrobial susceptibility pattern of clinical isolates of Pseudomonas aeruginosa isolated from patients of lower respiratory tract infections. SpringerPlus 2012, 1, 70. [Google Scholar] [CrossRef] [PubMed]
- Collins, C.H.; Lyne, P.M.; Grange, J.M.; Falkinham, J.O., III. Collins and Lyne’s Microbiological Methods, 8th ed.; Arnolds: London, UK, 2004. [Google Scholar]
- Forbes, B.; Sahm, D.; Weissfeld, A. Bailey and Scott’s Diagnostic Microbiology, 12th ed.; Mosby Elsevier: St. Louis, MO, USA, 2007. [Google Scholar]
- Basit, S.A.; Juayang, A.C. Outcomes-Based Education Manual in Clinical Bacteriology, 1st ed.; APD Educational Publishing House: Ermita, Manila, Philippines, 2016. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk Susceptibility Tests; CLSI: Wayne, PA, USA, 2012. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty Fifth Informational Supplement M100-S23; CLSI: Wayne, PA, USA, 2015. [Google Scholar]
- Polsfuss, S.; Bloemberg, G.; Giger, J.; Meyer, V.; Bottger, E.; Hombach, M. Practical approach for reliable detection of AmpC β-lactamase-producing Enterobacteriaceae. J. Clin. Microbiol. 2011, 49, 2798–2803. [Google Scholar] [CrossRef] [PubMed]
- Chika, E.; Charles, E.; Ifeanyichukwu, I.; Chigozie, U.; Chika, E.; Carissa, D.; Michael, A. Phenotypic detection of AmpC β-lactamase among anal Pseudomonas aeruginosa isolates in a Nigerian abattoir. Arch. Clin. Microbiol. 2016, 7, 2. [Google Scholar]
- Krumperman, P. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [PubMed]
- Davis, R.; Brown, P. Multiple antibiotic resistance index, fitness and virulence potential in respiratory Pseudomonas aeruginosa from Jamaica. J. Med. Microbiol. 2016, 65, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, G.A. AmpC β-lactamases. Clin. Microbiol. Rev. 2012, 22, 161–182. [Google Scholar] [CrossRef] [PubMed]
- Sedighi, M.; Safiri, S.; Pirouzi, S.; Jayasinghe, H.; Sepidarkish, M.; Fouladseresht, H. Detection and determination of the antibiotic resistance patterns in Pseudomonas aeruginosa strains isolated from clinical specimens in hospitals of Isfahan, Iran, 2012. Scimetr 2015, 3, e21133. [Google Scholar] [CrossRef]
- Tortora, G.; Funke, B.; Case, C. Microbiology: An Introduction, 6th ed.; Benjamin Cummings: California, CA, USA, 1998. [Google Scholar]
- Liu, Q.; Li, X.; Li, W.; Du, X.; He, J.-Q.; Tao, C.; Feng, Y. Influence of carbapenem resistance on mortality of patients with Pseudomonas aeruginosa infection: A meta-analysis. Sci. Rep. 2015, 5, 11715. [Google Scholar] [CrossRef] [PubMed]
- Thomson, K.S. Extended-spectrum-β-lactamase, AmpC, and carbapenemase issues. J. Clin. Microbiol. 2010, 48, 1019–1025. [Google Scholar] [CrossRef] [PubMed]
- Scheld, W.M.; Grayson, M.L.; Hughes, J.M. Infections in Long-Term Care Facilities in Emerging Infections, 9th ed.; ASM Press: New Washington, DC, USA, 2010; pp. 287–288. [Google Scholar]
- Yayan, J.; Ghebremedhin, B.; Rasche, K. Antibiotic resistance of Pseudomonas aeruginosa in pneumonia at a single university hospital center in Germany over a 10-year period. PLoS ONE 2015, 10, e0139836. [Google Scholar] [CrossRef] [PubMed]
- Berglund, B. Environmental dissemination of antibiotic resistance genes and correlation to anthropogenic contamination with antibiotics. J. Infect. Ecol. Epidemiol. 2015, 5, 28564. [Google Scholar] [CrossRef] [PubMed]
- Antimicrobial Resistance Surveillance Reference Laboratory. Antimicrobial Resistance Surveillance Program 2015 Data Summary Report; Research Institute for Tropical Medicine, Department of Health: Manila, Philippines, 2015.
- Pathmanathan, S.G.; Samat, N.A.; Mohamed, R. Antimicrobial susceptibility of clinical isolates of Pseudomonas aeruginosa from a Malaysian hospital. MJMS 2009, 16, 27–32. [Google Scholar] [PubMed]
- Juayang, A.; Maestral, D.; de los Reyes, G.; Acosido, M.A.; Gallega, C. Review on the antimicrobial resistance of pathogens from tracheal and endotracheal aspirates of patients with clinical manifestations of pneumonia in Bacolod City in 2013. Int. J. Bacteriol. 2015, 942509. [Google Scholar] [CrossRef] [PubMed]
- Kotra, L.; Haddad, J.; Mobashery, S. Aminoglycosides: Perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicrob. Agents Chemother. 2000, 44, 3249–3256. [Google Scholar] [CrossRef] [PubMed]
- Micek, S.; Lloyd, A.; Ritchie, D.; Reichley, R.; Fraser, V.; Kollef, M. Pseudomonas aeruginosa bloodstream infection: Importance of appropriate initial antimicrobial treatment. Antimicrob. Agents Chemother. 2005, 49, 1306–1311. [Google Scholar] [CrossRef] [PubMed]
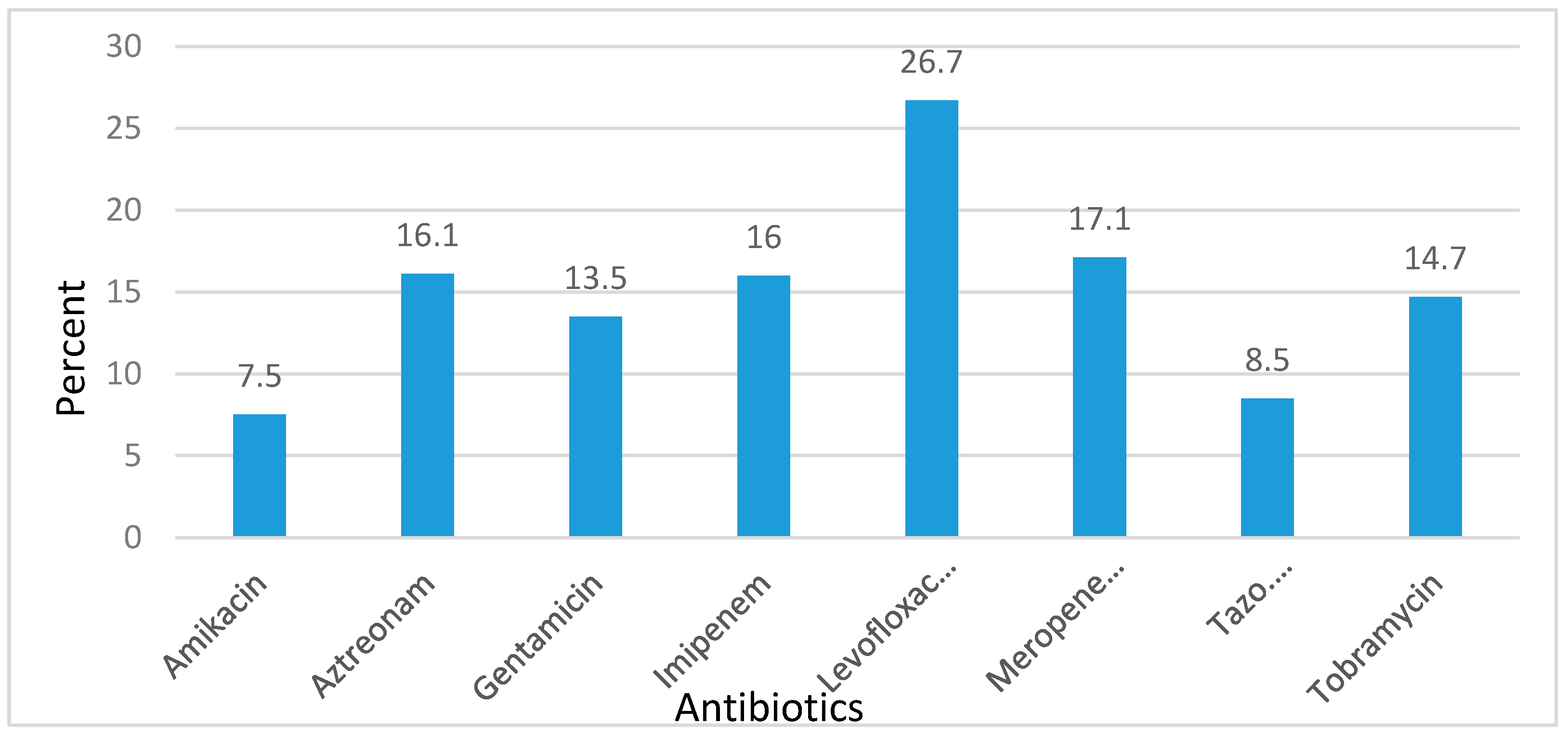
| Antibiotics | 2011 (n = 102) | 2012 (n = 68) | 2013 (n = 184) | 2014 (n = 142) | 2015 (n = 150) |
|---|---|---|---|---|---|
| Amikacin | 8 | 12 | 11 | 4 | 5 |
| Aztreonam | 16 | 15 | 22 | 18 | 9 |
| Gentamicin | 20 | 12 | 16 | 11 | 9 |
| Imipenem | 13 | 10 | 18 | 29 | 8 |
| Levofloxacin | 36 | 29 | 23 | 44 | 9 |
| Meropenem | 18 | 27 | 21 | 21 | 8 |
| Piperacillin/tazobactam | 5 | 8 | 8 | 11 | 8 |
| Tobramycin | 19 | 10 | 16 | 20 | 8 |
| Respiratory (n = 394) | Urinary (n = 150) | Blood and CSF (n = 25) | Wounds and Abscess (n = 64) | Transudates and Exudates (n = 13) | |
|---|---|---|---|---|---|
| Amikacin | 6.7 | 6.7 | 0 | 8.5 | 0 |
| Aztreonam | 17.8 | 12.9 | 6.2 | 8.7 | 53.8 |
| Cefoxitin | 96 | 95.1 | 100 | 100 | 100 |
| Gentamicin | 13.8 | 15.8 | 28.6 | 9.3 | 0 |
| Imipenem | 15.1 | 11.5 | 18.8 | 6.1 | 53.8 |
| Levofloxacin | 25.7 | 3.6 | 6.7 | 12.1 | 0 |
| Meropenem | 19.3 | 13.5 | 5.9 | 4.4 | 53.8 |
| Piperacillin/tazobactam | 8.3 | 9 | 0 | 2.4 | 53.8 |
| Tobramycin | 12.1 | 18 | 21.4 | 7.5 | 0 |
| MAR Index | Frequency Among Isolates |
|---|---|
| 0 | 271 |
| 0.1 | 136 |
| 0.2 | 79 |
| 0.3 | 41 |
| 0.4 | 32 |
| 0.5 | 29 |
| 0.6 | 28 |
| 0.7 | 8 |
| 0.8 | 3 |
| 0.9 | 4 |
| 1.0 | 15 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Juayang, A.C.; Lim, J.P.T.; Bonifacio, A.F.V.; Lambot, A.V.L.; Millan, S.M.; Sevilla, V.Z.J.N.; Sy, J.K.T.; Villanueva, P.J.; Grajales, C.P.; Gallega, C.T. Five-Year Antimicrobial Susceptibility of Pseudomonas aeruginosa from a Local Tertiary Hospital in Bacolod City, Philippines. Trop. Med. Infect. Dis. 2017, 2, 28. https://doi.org/10.3390/tropicalmed2030028
Juayang AC, Lim JPT, Bonifacio AFV, Lambot AVL, Millan SM, Sevilla VZJN, Sy JKT, Villanueva PJ, Grajales CP, Gallega CT. Five-Year Antimicrobial Susceptibility of Pseudomonas aeruginosa from a Local Tertiary Hospital in Bacolod City, Philippines. Tropical Medicine and Infectious Disease. 2017; 2(3):28. https://doi.org/10.3390/tropicalmed2030028
Chicago/Turabian StyleJuayang, Alain C., Joseph Peter T. Lim, Ann Francis V. Bonifacio, Alaica Victoria L. Lambot, Sean Maybelle Millan, Vic Zyrus Jeriko N. Sevilla, Julien Kate T. Sy, Paul John Villanueva, Carmina P. Grajales, and Christine T. Gallega. 2017. "Five-Year Antimicrobial Susceptibility of Pseudomonas aeruginosa from a Local Tertiary Hospital in Bacolod City, Philippines" Tropical Medicine and Infectious Disease 2, no. 3: 28. https://doi.org/10.3390/tropicalmed2030028





