Small RNAs in Circulating Exosomes of Cancer Patients: A Minireview
Abstract
:1. Small Non-Coding RNAs
2. MicroRNA Functions and Roles in Cancer Development
3. Extracellular Vesicles Contain, Transport and Deliver small RNAs
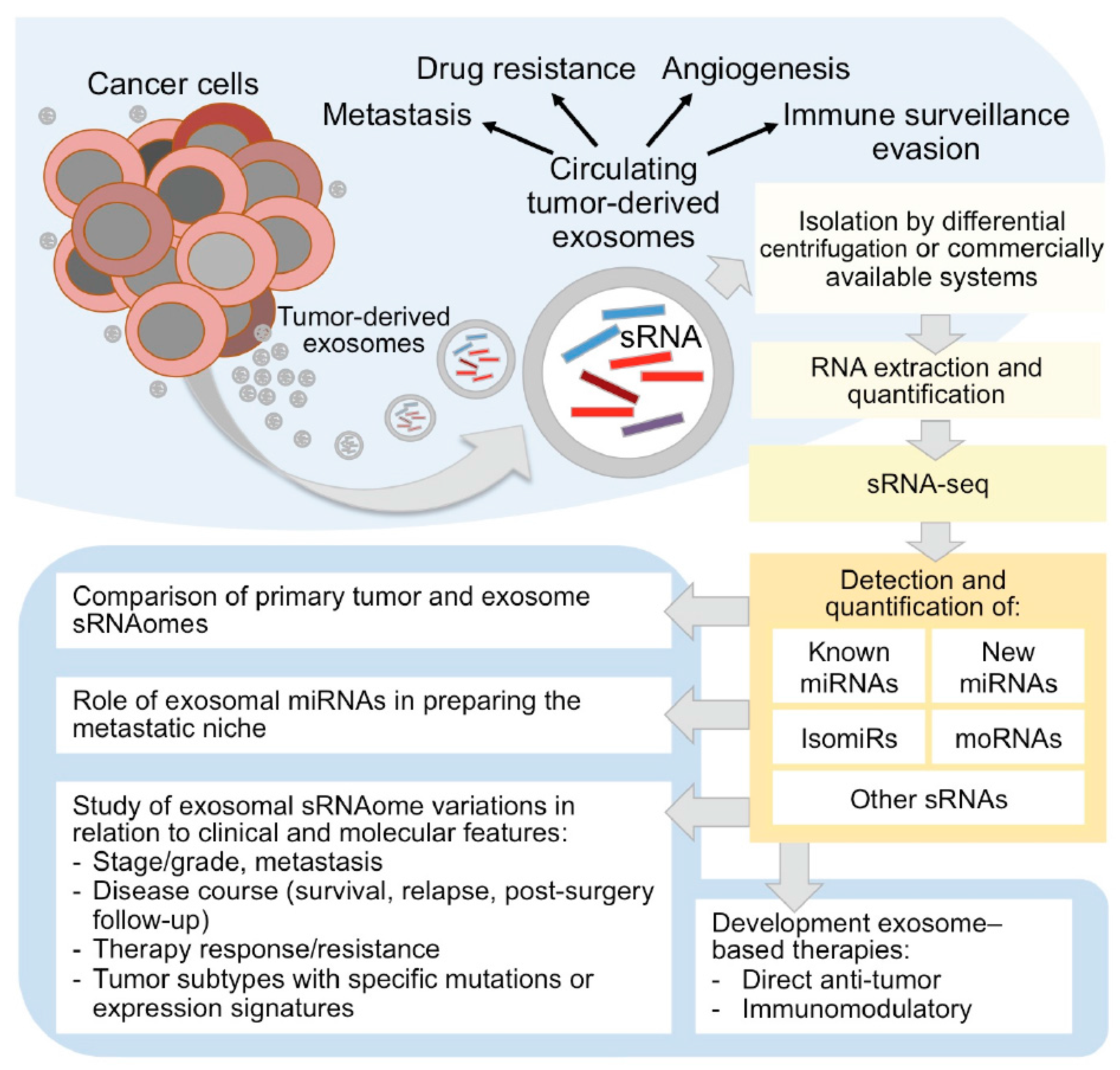
4. Exosomes Roles in Cancer Development and Progression and Relevance as Biomarkers
5. Development of Exosome-Based Therapies
6. Experimental and Computational Procedures to Study Circulating Exosomal small RNAs by RNA-Seq
6.1. Exosome RNA Isolation and Sequencing
6.2. Bioinformatic Analysis of Exosomal sRNA-Seq Data
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Piatek, M.J.; Werner, A. Endogenous siRNAs: Regulators of internal affairs. Biochem. Soc. Trans. 2014, 42, 1174–1179. [Google Scholar] [CrossRef] [PubMed]
- Le Thomas, A.; Tóth, K.F.; Aravin, A.A. To be or not to be a piRNA: Genomic origin and processing of piRNAs. Genome Biol. 2014, 15, 204. [Google Scholar] [CrossRef] [PubMed]
- Moyano, M.; Stefani, G. piRNA involvement in genome stability and human cancer. J. Hematol. Oncol. 2015, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Eichhorn, S.W.; Guo, H.; McGeary, S.E.; Rodriguez-Mias, R.A.; Shin, C.; Baek, D.; Hsu, S.-H.; Ghoshal, K.; Villén, J.; Bartel, D.P. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol. Cell 2014, 56, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Volpe, T.A. Regulation of Heterochromatic Silencing and Histone H3 Lysine-9 Methylation by RNAi. Science 2002, 297, 1833–1837. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Zhang, B.; Wu, T.; Skogerbo, G.; Zhu, X.; Guo, X.; He, S.; Chen, R. Transcriptional inhibition of Hoxd4 expression by noncoding RNAs in human breast cancer cells. BMC Mol. Biol. 2009, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Ghildiyal, M.; Zamore, P.D. Small silencing RNAs: An expanding universe. Nat. Rev. Genet. 2009, 10, 94–108. [Google Scholar] [CrossRef] [PubMed]
- Pasquinelli, A.E. MicroRNAs and their targets: Recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 2012, 13, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Narry Kim, V. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [PubMed]
- Guglielmelli, P.; Bisognin, A.; Saccoman, C.; Mannarelli, C.; Coppe, A.; Vannucchi, A.M.; Bortoluzzi, S. Small RNA Sequencing Uncovers New miRNAs and moRNAs Differentially Expressed in Normal and Primary Myelofibrosis CD34+ Cells. PLoS ONE 2015, 10, e0140445. [Google Scholar] [CrossRef] [PubMed]
- Bortoluzzi, S.; Bisognin, A.; Biasiolo, M.; Guglielmelli, P.; Biamonte, F.; Norfo, R.; Manfredini, R.; Vannucchi, A.M.; AGIMM (Associazione Italiana per la Ricerca sul Cancro–Gruppo Italiano Malattie Mieloproliferative) Investigators. Characterization and discovery of novel miRNAs and moRNAs in JAK2V617F-mutated SET2 cells. Blood 2012, 119, e120–e130. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.C.; Dibb, N. IsomiRs have functional importance. Malays. J. Pathol. 2015, 37, 73–81. [Google Scholar] [PubMed]
- Bortoluzzi, S.; Biasiolo, M.; Bisognin, A. MicroRNA-offset RNAs (moRNAs): By-product spectators or functional players? Trends Mol. Med. 2011, 17, 473–474. [Google Scholar] [CrossRef] [PubMed]
- Abdelfattah, A.M.; Park, C.; Choi, M.Y. Update on non-canonical microRNAs. Biomol. Concepts 2014, 5, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Maute, R.L.; Dalla-Favera, R.; Basso, K. RNAs with multiple personalities. Wiley Interdiscip. Rev. RNA 2014, 5, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Curtis, H.J.; Sibley, C.R.; Wood, M.J.A. Mirtrons, an emerging class of atypical miRNA. Wiley Interdiscip. Rev. RNA 2012, 3, 617–632. [Google Scholar] [CrossRef] [PubMed]
- Scott, M.S.; Ono, M. From snoRNA to miRNA: Dual function regulatory non-coding RNAs. Biochimie 2011, 93, 1987–1992. [Google Scholar] [CrossRef] [PubMed]
- Falaleeva, M.; Stamm, S. Processing of snoRNAs as a new source of regulatory non-coding RNAs. Bioessays 2012, 35, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P.; Ivanov, P. tRNA fragments in human health and disease. FEBS Lett. 2014, 588, 4297–4304. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, T.; Suresh, P.S.; Tsutsumi, R. tRFs: miRNAs in disguise. Gene 2016, 579, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Langenberger, D.; Çakir, M.V.; Hoffmann, S.; Stadler, P.F. Dicer-processed small RNAs: Rules and exceptions. J. Exp. Zool. B Mol. Dev. Evol. 2013, 320, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Persson, H.; Kvist, A.; Vallon-Christersson, J.; Medstrand, P.; Borg, A.; Rovira, C. The non-coding RNA of the multidrug resistance-linked vault particle encodes multiple regulatory small RNAs. Nat. Cell Biol. 2009, 11, 1268–1271. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, F.E.; Hall, A.E.; Csorba, T.; Turnbull, C.; Dalmay, T. Biogenesis of Y RNA-derived small RNAs is independent of the microRNA pathway. FEBS Lett. 2012, 586, 1226–1230. [Google Scholar] [CrossRef] [PubMed]
- Kapralova, K.H.; Franzdóttir, S.R.; Jónsson, H.; Snorrason, S.S.; Jónsson, Z.O. Patterns of miRNA expression in Arctic charr development. PLoS ONE 2014, 9, e106084. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, S.; Yan, F.; Liu, X.; Jiang, R.; Han, R.; Li, Z.; Li, G.; Tian, Y.; Kang, X.; et al. Effect of polymorphism within miRNA-1606 gene on growth and carcass traits in chicken. Gene 2015, 566, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-Z.; Li, L.; Lodish, H.F.; Bartel, D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004, 303, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Qin, Y.-W.; Brewer, G.; Jing, Q. MicroRNA degradation and turnover: Regulating the regulators. Wiley Interdiscip. Rev. RNA 2012, 3, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Bonizzato, A.; Gaffo, E.; Te Kronnie, G.; Bortoluzzi, S. CircRNAs in hematopoiesis and hematological malignancies. Blood Cancer J. 2016, 6, e483. [Google Scholar] [CrossRef] [PubMed]
- Markopoulos, G.S.; Roupakia, E.; Tokamani, M.; Chavdoula, E.; Hatziapostolou, M.; Polytarchou, C.; Marcu, K.B.; Papavassiliou, A.G.; Sandaltzopoulos, R.; Kolettas, E. A step-by-step microRNA guide to cancer development and metastasis. Cell. Oncol. 2017, 40, 303–339. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Croce, C.M. MicroRNA-cancer connection: The beginning of a new tale. Cancer Res. 2006, 66, 7390–7394. [Google Scholar] [CrossRef] [PubMed]
- Inui, M.; Martello, G.; Piccolo, S. MicroRNA control of signal transduction. Nat. Rev. Mol. Cell Biol. 2010, 11, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Bisognin, A.; Sales, G.; Coppe, A.; Bortoluzzi, S.; Romualdi, C. MAGIA2: From miRNA and genes expression data integrative analysis to microRNA-transcription factor mixed regulatory circuits (2012 update). Nucleic Acids Res. 2012, 40, W13–W21. [Google Scholar] [CrossRef] [PubMed]
- Calura, E.; Bisognin, A.; Manzoni, M.; Todoerti, K.; Taiana, E.; Sales, G.; Morgan, G.J.; Tonon, G.; Amodio, N.; Tassone, P.; et al. Disentangling the microRNA regulatory milieu in multiple myeloma: Integrative genomics analysis outlines mixed miRNA-TF circuits and pathway-derived networks modulated in t(4;14) patients. Oncotarget 2016, 7, 2367–2378. [Google Scholar] [CrossRef] [PubMed]
- Calura, E.; Pizzini, S.; Bisognin, A.; Coppe, A.; Sales, G.; Gaffo, E.; Fanelli, T.; Mannarelli, C.; Zini, R.; Norfo, R.; et al. A data-driven network model of primary myelofibrosis: Transcriptional and post-transcriptional alterations in CD34+ cells. Blood Cancer J. 2016, 6, e439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pizzini, S.; Bisognin, A.; Mandruzzato, S.; Biasiolo, M.; Facciolli, A.; Perilli, L.; Rossi, E.; Esposito, G.; Rugge, M.; Pilati, P.; et al. Impact of microRNAs on regulatory networks and pathways in human colorectal carcinogenesis and development of metastasis. BMC Genomics 2013, 14, 589. [Google Scholar] [CrossRef] [PubMed]
- Bonci, D.; Coppola, V.; Patrizii, M.; Addario, A.; Cannistraci, A.; Francescangeli, F.; Pecci, R.; Muto, G.; Collura, D.; Bedini, R.; et al. A microRNA code for prostate cancer metastasis. Oncogene 2016, 35, 1180–1192. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Weinberg, R.A. Micromanagers of malignancy: Role of microRNAs in regulating metastasis. Trends Genet. 2008, 24, 448–456. [Google Scholar] [CrossRef] [PubMed]
- Sonda, N.; Simonato, F.; Peranzoni, E.; Calì, B.; Bortoluzzi, S.; Bisognin, A.; Wang, E.; Marincola, F.M.; Naldini, L.; Gentner, B.; et al. miR-142-3p prevents macrophage differentiation during cancer-induced myelopoiesis. Immunity 2013, 38, 1236–1249. [Google Scholar] [CrossRef] [PubMed]
- Noman, M.Z.; Janji, B.; Hu, S.; Wu, J.C.; Martelli, F.; Bronte, V.; Chouaib, S. Tumor-Promoting Effects of Myeloid-Derived Suppressor Cells Are Potentiated by Hypoxia-Induced Expression of miR-210. Cancer Res. 2015, 75, 3771–3787. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liu, Q.; Mi, S.; Liang, X.; Zhang, Z.; Su, X.; Liu, J.; Chen, Y.; Wang, M.; Zhang, Y.; et al. Both miR-17-5p and miR-20a alleviate suppressive potential of myeloid-derived suppressor cells by modulating STAT3 expression. J. Immunol. 2011, 186, 4716–4724. [Google Scholar] [CrossRef] [PubMed]
- Tran, M.N.; Choi, W.; Wszolek, M.F.; Navai, N.; Lee, I.-L.C.; Nitti, G.; Wen, S.; Flores, E.R.; Siefker-Radtke, A.; Czerniak, B.; et al. The p63 protein isoform ΔNp63α inhibits epithelial-mesenchymal transition in human bladder cancer cells: Role of MIR-205. J. Biol. Chem. 2013, 288, 3275–3288. [Google Scholar] [CrossRef] [PubMed]
- Perilli, L.; Vicentini, C.; Agostini, M.; Pizzini, S.; Pizzi, M.; D’Angelo, E.; Bortoluzzi, S.; Mandruzzato, S.; Mammano, E.; Rugge, M.; et al. Circulating miR-182 is a biomarker of colorectal adenocarcinoma progression. Oncotarget 2014, 5, 6611–6619. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Fabbri, M.; Calin, G.A. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat. Rev. Drug Discov. 2013, 12, 847–865. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Aalberts, M.; van Dissel-Emiliani, F.M.F.; van Adrichem, N.P.H.; van Wijnen, M.; Wauben, M.H.M.; Stout, T.A.E.; Stoorvogel, W. Identification of distinct populations of prostasomes that differentially express prostate stem cell antigen, annexin A1, and GLIPR2 in humans. Biol. Reprod. 2012, 86, 82. [Google Scholar] [CrossRef] [PubMed]
- Caby, M.-P.; Lankar, D.; Vincendeau-Scherrer, C.; Raposo, G.; Bonnerot, C. Exosomal-like vesicles are present in human blood plasma. Int. Immunol. 2005, 17, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Pisitkun, T.; Shen, R.-F.; Knepper, M.A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. USA 2004, 101, 13368–13373. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, Y.; Miura, Y.; Harazono, A.; Kanai-Azuma, M.; Akimoto, Y.; Kawakami, H.; Yamaguchi, T.; Toda, T.; Endo, T.; Tsubuki, M.; et al. Proteomic analysis of two types of exosomes in human whole saliva. Biol. Pharm. Bull. 2011, 34, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Admyre, C.; Johansson, S.M.; Qazi, K.R.; Filén, J.-J.; Lahesmaa, R.; Norman, M.; Neve, E.P.A.; Scheynius, A.; Gabrielsson, S. Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 2007, 179, 1969–1978. [Google Scholar] [CrossRef] [PubMed]
- Asea, A.; Jean-Pierre, C.; Kaur, P.; Rao, P.; Linhares, I.M.; Skupski, D.; Witkin, S.S. Heat shock protein-containing exosomes in mid-trimester amniotic fluids. J. Reprod. Immunol. 2008, 79, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Andre, F.; Schartz, N.E.C.; Movassagh, M.; Flament, C.; Pautier, P.; Morice, P.; Pomel, C.; Lhomme, C.; Escudier, B.; Le Chevalier, T.; et al. Malignant effusions and immunogenic tumour-derived exosomes. Lancet 2002, 360, 295–305. [Google Scholar] [CrossRef]
- Vella, L.J.; Sharples, R.A.; Lawson, V.A.; Masters, C.L.; Cappai, R.; Hill, A.F. Packaging of prions into exosomes is associated with a novel pathway of PrP processing. J. Pathol. 2007, 211, 582–590. [Google Scholar] [CrossRef] [PubMed]
- Milani, G.; Lana, T.; Bresolin, S.; Aveic, S.; Pastò, A.; Frasson, C.; Te Kronnie, G. Expression Profiling of Circulating Microvesicles Reveals Intercellular Transmission of Oncogenic Pathways. Mol. Cancer Res. 2017, 15, 683–695. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, F.A.; Lécuyer, E. Small Luggage for a Long Journey: Transfer of Vesicle-Enclosed Small RNA in Interspecies Communication. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Xiang, L.; He, L.; Yang, G.; Zheng, J.; Wang, C.; Zhang, Y.; Wang, S.; Zhou, Y.; Sheu, T.-J.; et al. Exosomes Mediate Epithelium–Mesenchyme Crosstalk in Organ Development. ACS Nano 2017. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-W.; Li, S.; Lin, J.D. The Micro-Managing Fat: Exosomes as a New Messenger. Trends Endocrinol. Metab. 2017, 28, 541–542. [Google Scholar] [CrossRef] [PubMed]
- Skokos, D.; Goubran-Botros, H.; Roa, M.; Mécheri, S. Immunoregulatory properties of mast cell-derived exosomes. Mol. Immunol. 2002, 38, 1359–1362. [Google Scholar] [PubMed]
- Huang, X.; Yuan, T.; Tschannen, M.; Sun, Z.; Jacob, H.; Du, M.; Liang, M.; Dittmar, R.L.; Liu, Y.; Liang, M.; et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics 2013, 14, 319. [Google Scholar] [CrossRef] [PubMed]
- Sato-Kuwabara, Y.; Melo, S.A.; Soares, F.A.; Calin, G.A. The fusion of two worlds: Non-coding RNAs and extracellular vesicles-diagnostic and therapeutic implications. Int. J. Oncol. 2015, 46, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D.; Homesley, H.D.; Doellgast, G.J. “Membrane-associated” immunoglobulins in cyst and ascites fluids of ovarian cancer patients. Am. J. Reprod. Immunol. 1983, 3, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.F.; Li, S.; Chin, A.R.; et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 2015, 17, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Sun, Y.; Peng, J.; Liao, H.; Gao, H.; Guo, Y.; Guo, L. Overexpression of secretagogin inhibits cell apoptosis and induces chemoresistance in small cell lung cancer under the regulation of miR-494. Oncotarget 2014, 5, 7760–7775. [Google Scholar] [CrossRef] [PubMed]
- Peinado, H.; Alečković, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; García-Santos, G.; Ghajar, C.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szajnik, M.; Czystowska, M.; Szczepanski, M.J.; Mandapathil, M.; Whiteside, T.L. Tumor-derived microvesicles induce, expand and up-regulate biological activities of human regulatory T cells (Treg). PLoS ONE 2010, 5, e11469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallach, S.; Jantus-Lewintre, E.; Calabuig-Fariñas, S.; Montaner, D.; Alonso, S.; Sirera, R.; Blasco, A.; Usó, M.; Guijarro, R.; Martorell, M.; et al. MicroRNA profiling associated with non-small cell lung cancer: Next generation sequencing detection, experimental validation, and prognostic value. Oncotarget 2017. [Google Scholar] [CrossRef] [PubMed]
- Que, R.; Ding, G.; Chen, J.; Cao, L. Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma. World J. Surg. Oncol. 2013, 11, 219. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, C.; Rani, S.; O’Driscoll, L. miR-34a is an intracellular and exosomal predictive biomarker for response to docetaxel with clinical relevance to prostate cancer progression. Prostate 2014, 74, 1320–1334. [Google Scholar] [CrossRef] [PubMed]
- Lunavat, T.R.; Cheng, L.; Einarsdottir, B.O.; Olofsson Bagge, R.; Veppil Muralidharan, S.; Sharples, R.A.; Lässer, C.; Gho, Y.S.; Hill, A.F.; Nilsson, J.A.; et al. BRAF(V600) inhibition alters the microRNA cargo in the vesicular secretome of malignant melanoma cells. Proc. Natl. Acad. Sci. USA 2017, 114, E5930–E5939. [Google Scholar] [CrossRef] [PubMed]
- van Eijndhoven, M.A.J.; Zijlstra, J.M.; Groenewegen, N.J.; Drees, E.E.E.; van Niele, S.; Baglio, S.R.; Koppers-Lalic, D.; van der Voorn, H.; Libregts, S.F.W.M.; Wauben, M.H.M.; et al. Plasma vesicle miRNAs for therapy response monitoring in Hodgkin lymphoma patients. JCI Insight 2016, 1, e89631. [Google Scholar] [CrossRef] [PubMed]
- Challagundla, K.B.; Wise, P.M.; Neviani, P.; Chava, H.; Murtadha, M.; Xu, T.; Kennedy, R.; Ivan, C.; Zhang, X.; Vannini, I.; et al. Exosome-mediated transfer of microRNAs within the tumor microenvironment and neuroblastoma resistance to chemotherapy. J. Natl. Cancer Inst. 2015, 107. [Google Scholar] [CrossRef] [PubMed]
- Rana, S.; Malinowska, K.; Zöller, M. Exosomal tumor microRNA modulates premetastatic organ cells. Neoplasia 2013, 15, 281–295. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, C.A.; Andahur, E.I.; Valenzuela, R.; Castellón, E.A.; Fullá, J.A.; Ramos, C.G.; Triviño, J.C. Exosomes from bulk and stem cells from human prostate cancer have a differential microRNA content that contributes cooperatively over local and pre-metastatic niche. Oncotarget 2016, 7, 3993–4008. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, C.; Wang, S.; Wang, Z.; Jiang, J.; Wang, W.; Li, X.; Chen, J.; Liu, K.; Li, C.; et al. Exosomes Derived from Hypoxic Oral Squamous Cell Carcinoma Cells Deliver miR-21 to Normoxic Cells to Elicit a Prometastatic Phenotype. Cancer Res. 2016, 76, 1770–1780. [Google Scholar] [CrossRef] [PubMed]
- Mittelbrunn, M.; Gutiérrez-Vázquez, C.; Villarroya-Beltri, C.; González, S.; Sánchez-Cabo, F.; González, M.Á.; Bernad, A.; Sánchez-Madrid, F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2011, 2, 282. [Google Scholar] [CrossRef] [PubMed]
- Shimbo, K.; Miyaki, S.; Ishitobi, H.; Kato, Y.; Kubo, T.; Shimose, S.; Ochi, M. Exosome-formed synthetic microRNA-143 is transferred to osteosarcoma cells and inhibits their migration. Biochem. Biophys. Res. Commun. 2014, 445, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Munoz, J.L.; Bliss, S.A.; Greco, S.J.; Ramkissoon, S.H.; Ligon, K.L.; Rameshwar, P. Delivery of Functional Anti-miR-9 by Mesenchymal Stem Cell-derived Exosomes to Glioblastoma Multiforme Cells Conferred Chemosensitivity. Mol. Ther. Nucleic Acids 2013, 2, e126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, A.F.; Michiel Pegtel, D.; Lambertz, U.; Leonardi, T.; O’Driscoll, L.; Pluchino, S.; Ter-Ovanesyan, D.; Hoen, E.N.M.N.-‘t. ISEV position paper: Extracellular vesicle RNA analysis and bioinformatics. J. Extracell. Vesicles 2013, 2, 22859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reiner, A.T.; Witwer, K.W.; van Balkom, B.W.M.; de Beer, J.; Brodie, C.; Corteling, R.L.; Gabrielsson, S.; Gimona, M.; Ibrahim, A.G.; de Kleijn, D.; et al. Concise Review: Developing Best-Practice Models for the Therapeutic Use of Extracellular Vesicles. Stem Cells Transl. Med. 2017, 6, 1730–1739. [Google Scholar] [CrossRef] [PubMed]
- Caivano, A.; La Rocca, F.; Laurenzana, I.; Trino, S.; De Luca, L.; Lamorte, D.; Del Vecchio, L.; Musto, P. Extracellular Vesicles in Hematological Malignancies: From Biology to Therapy. Int. J. Mol. Sci. 2017, 18, 1183. [Google Scholar] [CrossRef] [PubMed]
- Witwer, K.W.; Buzás, E.I.; Bemis, L.T.; Bora, A.; Lässer, C.; Lötvall, J.; Nolte-’t Hoen, E.N.; Piper, M.G.; Sivaraman, S.; Skog, J.; et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles 2013, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rekker, K.; Saare, M.; Roost, A.M.; Kubo, A.-L.; Zarovni, N.; Chiesi, A.; Salumets, A.; Peters, M. Comparison of serum exosome isolation methods for microRNA profiling. Clin. Biochem. 2014, 47, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Lane, R.E.; Korbie, D.; Anderson, W.; Vaidyanathan, R.; Trau, M. Analysis of exosome purification methods using a model liposome system and tunable-resistive pulse sensing. Sci. Rep. 2015, 5, 7639. [Google Scholar] [CrossRef] [PubMed]
- Enderle, D.; Spiel, A.; Coticchia, C.M.; Berghoff, E.; Mueller, R.; Schlumpberger, M.; Sprenger-Haussels, M.; Shaffer, J.M.; Lader, E.; Skog, J.; et al. Characterization of RNA from Exosomes and Other Extracellular Vesicles Isolated by a Novel Spin Column-Based Method. PLoS ONE 2015, 10, e0136133. [Google Scholar] [CrossRef] [PubMed]
- Yuan, T.; Huang, X.; Woodcock, M.; Du, M.; Dittmar, R.; Wang, Y.; Tsai, S.; Kohli, M.; Boardman, L.; Patel, T.; et al. Plasma extracellular RNA profiles in healthy and cancer patients. Sci. Rep. 2016, 6, 19413. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Roehr, J.T.; Dieterich, C.; Reinert, K. Flexbar 3.0—SIMD and multicore parallelization. Bioinformatics 2017. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinform. 2010. Chapter 11, Unit 11.7. [Google Scholar]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Gaffo, E.; Zambonelli, P.; Bisognin, A.; Bortoluzzi, S.; Davoli, R. miRNome of Italian Large White pig subcutaneous fat tissue: New miRNAs, isomiRs and moRNAs. Anim. Genet. 2014, 45, 685–698. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Gordon, W.; Du, S.; Zhang, C.; He, W.; Xi, L.; Mathur, S.; Agostino, M.; Paradis, T.; von Schack, D.; et al. QuickMIRSeq: A pipeline for quick and accurate quantification of both known miRNAs and isomiRs by jointly processing multiple samples from microRNA sequencing. BMC Bioinform. 2017, 18, 180. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Yu, J.; Liang, T.; Zou, Q. miR-isomiRExp: A web-server for the analysis of expression of miRNA at the miRNA/isomiR levels. Sci. Rep. 2016, 6, 23700. [Google Scholar] [CrossRef] [PubMed]
- Telonis, A.G.; Magee, R.; Loher, P.; Chervoneva, I.; Londin, E.; Rigoutsos, I. Knowledge about the presence or absence of miRNA isoforms (isomiRs) can successfully discriminate amongst 32 TCGA cancer types. Nucleic Acids Res. 2017, 45, 2973–2985. [Google Scholar] [CrossRef] [PubMed]
- Wallaert, A.; Van Loocke, W.; Hernandez, L.; Taghon, T.; Speleman, F.; Van Vlierberghe, P. Comprehensive miRNA expression profiling in human T-cell acute lymphoblastic leukemia by small RNA-sequencing. Sci. Rep. 2017, 7, 7901. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Liang, T.; Yu, J.; Zou, Q. A Comprehensive Analysis of miRNA/isomiR Expression with Gender Difference. PLoS ONE 2016, 11, e0154955. [Google Scholar] [CrossRef] [PubMed]
- Koppers-Lalic, D.; Hackenberg, M.; Bijnsdorp, I.V.; van Eijndhoven, M.A.J.; Sadek, P.; Sie, D.; Zini, N.; Middeldorp, J.M.; Ylstra, B.; de Menezes, R.X.; et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014, 8, 1649–1658. [Google Scholar] [CrossRef] [PubMed]
- Veneziano, D.; Di Bella, S.; Nigita, G.; Laganà, A.; Ferro, A.; Croce, C.M. Noncoding RNA: Current Deep Sequencing Data Analysis Approaches and Challenges. Hum. Mutat. 2016, 37, 1283–1298. [Google Scholar] [CrossRef] [PubMed]
- Abels, E.R.; Breakefield, X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell. Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Tosar, J.P.; Cayota, A.; Eitan, E.; Halushka, M.K.; Witwer, K.W. Ribonucleic artefacts: Are some extracellular RNA discoveries driven by cell culture medium components? J. Extracell. Vesicles 2017, 6, 1272832. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bortoluzzi, S.; Lovisa, F.; Gaffo, E.; Mussolin, L. Small RNAs in Circulating Exosomes of Cancer Patients: A Minireview. High-Throughput 2017, 6, 13. https://doi.org/10.3390/ht6040013
Bortoluzzi S, Lovisa F, Gaffo E, Mussolin L. Small RNAs in Circulating Exosomes of Cancer Patients: A Minireview. High-Throughput. 2017; 6(4):13. https://doi.org/10.3390/ht6040013
Chicago/Turabian StyleBortoluzzi, Stefania, Federica Lovisa, Enrico Gaffo, and Lara Mussolin. 2017. "Small RNAs in Circulating Exosomes of Cancer Patients: A Minireview" High-Throughput 6, no. 4: 13. https://doi.org/10.3390/ht6040013





