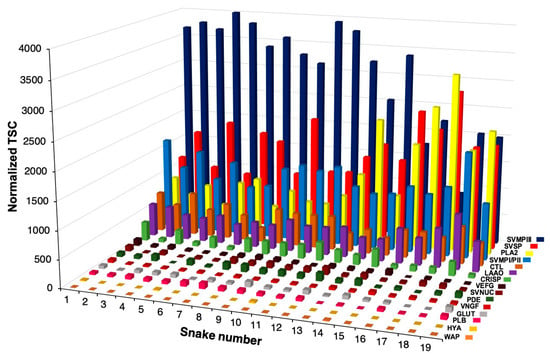
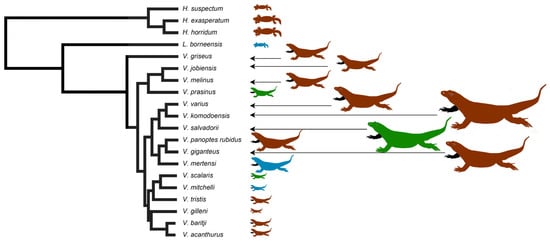
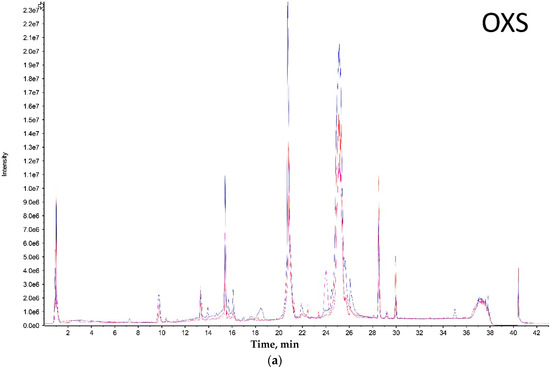
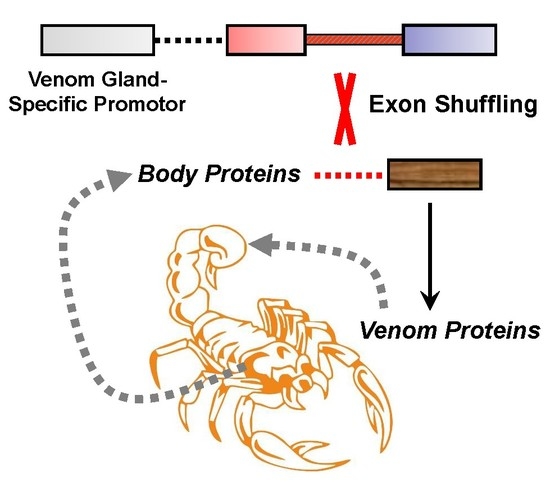
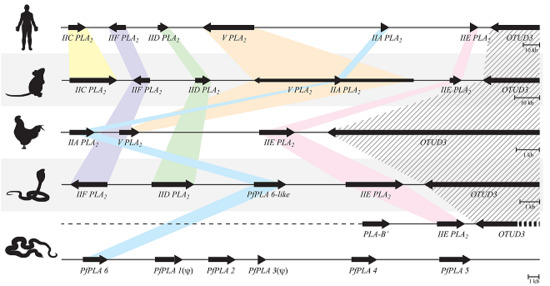
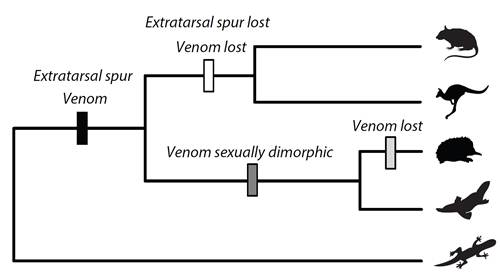
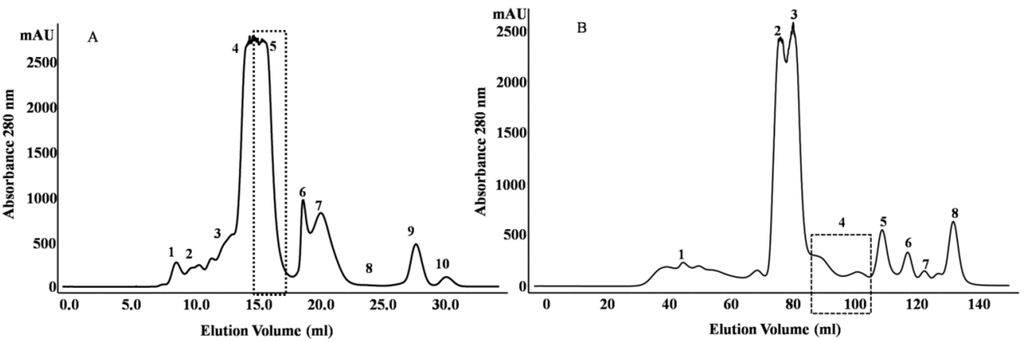
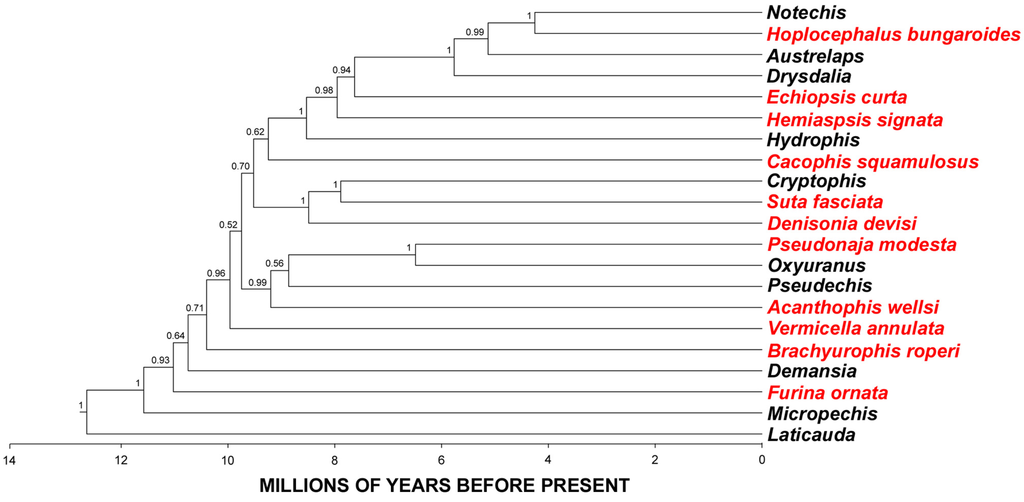
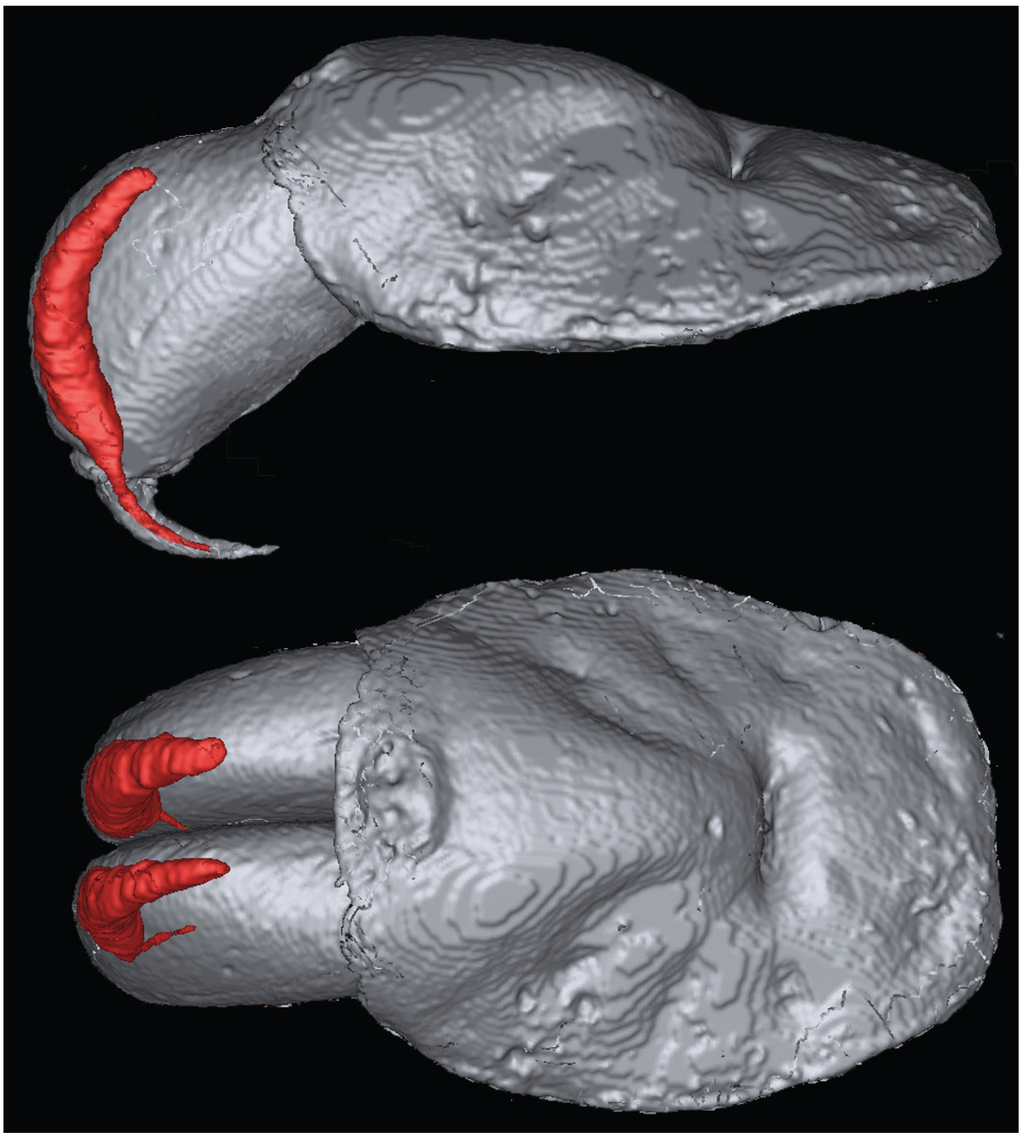
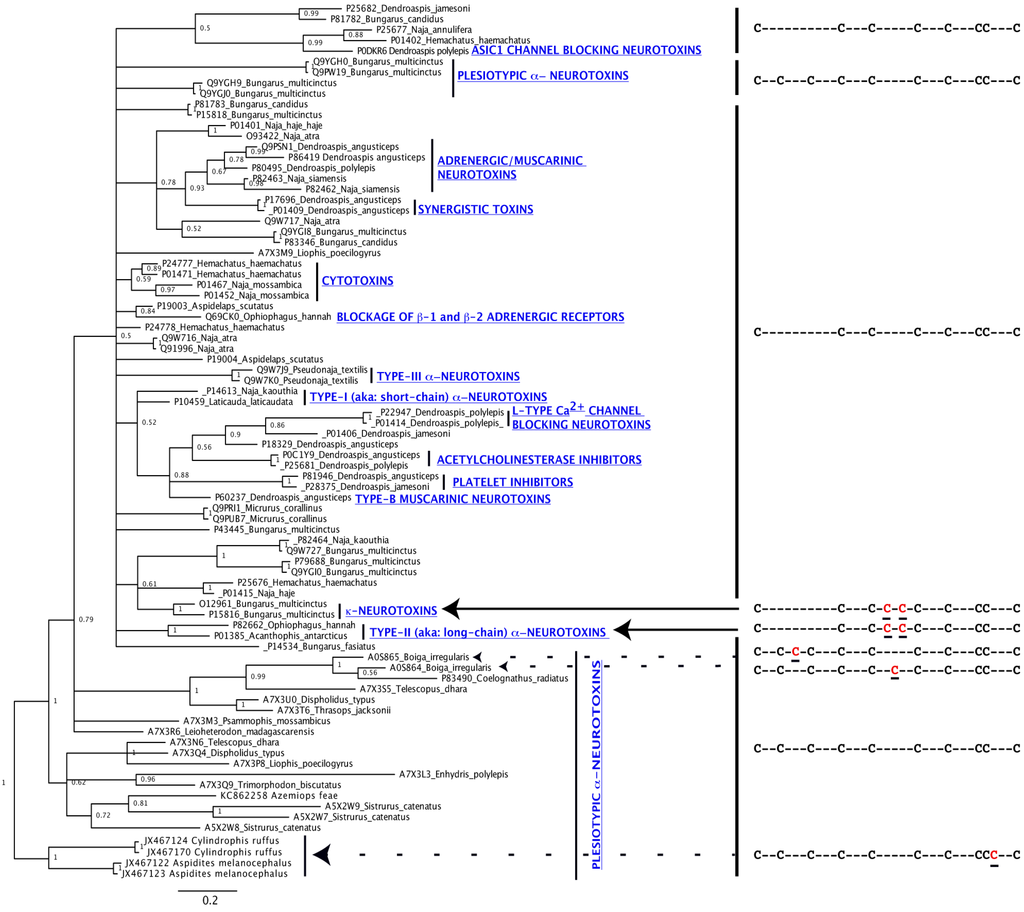
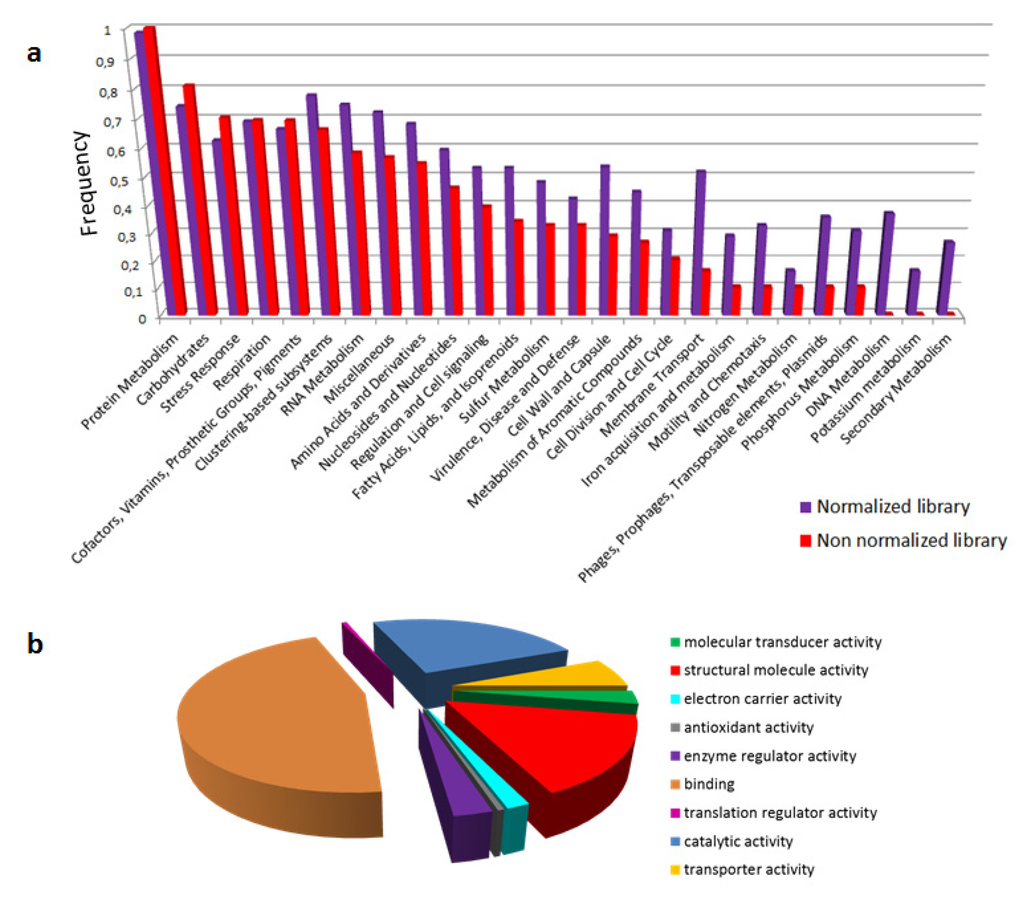
Evolution of Venom Systems
A topical collection in Toxins (ISSN 2072-6651). This collection belongs to the section "Animal Venoms".
Viewed by 241208Editor
Interests: venom molecular evolution; phylogenetics and structure–function relationships; toxins
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
Venom systems are key evolutionary innovations used for competitor deterrence, defence and predation. The evolution of venom has been the trigger for the explosive radiation of many orders of animals. This Special Issue will be composed of papers exploring origin and diversification of venom systems and their intricate relationship with changes in predatory ecology. Reviews for this collection are on an invitation-only basis. Please contact Professor Fry to discuss ideas for a review so as to avoid overlap with other reviews.
Prof. Dr. Bryan Fry
Guest Editor
Manuscript Submission Information
Manuscripts for the topical collection can be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. All papers will be peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on this website. The topical collection considers regular research articles, short communications and review articles. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page.
Please visit the Instructions for Authors page before submitting a manuscript. The article processing charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs).