The Histone Deacetylase Family: Structural Features and Application of Combined Computational Methods
Abstract
:1. Introduction
1.1. Classification of HDAC Family
1.2. HDAC Structure and Function


1.3. Mutation Effects on HDACs Biology
1.4. HDACs, HDACIs, Metabolism and Emerging Technologies like Omics
2. HDAC Inhibitors
- (1)
- the cap structure (Surface Recognition Domain), which typically constitutes a hydrophobic aromatic moiety that interfaces with the enzyme surface;
- (2)
- a Zn2+ binding group (ZBG), such as isohydroxamic acid, carboxylic acid, or benzamide, coordinating the Zn2+ ion at the enzyme catalytic center;
- (3)
- (1)
- Isohydroxamic acids, which encompass SAHA, Belinostat (PXD101), and Panobinostat (LBH589);
- (2)
- Benzamide derivatives, exemplified by Mocetinostat (MGCD0103) and Chidamide;
- (3)
- Cyclic peptides, represented by romidepsin (FK228) [138].
2.1. Isohydroxamic Acids
2.2. Benzamide Derivatives
2.3. Cyclic Peptides
2.4. Future Perspectives and PROTACs
3. Animal Research and Clinical Trials with HDAC Inhibitors
4. Computational Studies on HDACs
4.1. Molecular Modeling
4.2. Machine Learning
4.3. Limitations of Computational Techniques
4.4. Achieving Selectivity for Each HDAC Isoform
5. In Vitro Validations Using Cell Lines
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Zhang, Q.; Dai, Y.; Cai, Z.; Mou, L. HDAC Inhibitors: Novel Immunosuppressants for Allo- and Xeno-Transplantation. Chem. Sel. 2018, 3, 176–187. [Google Scholar] [CrossRef]
- Milazzo, G.; Mercatelli, D.; Di Muzio, G.; Triboli, L.; De Rosa, P.; Perini, G.; Giorgi, F.M. Histone Deacetylases (HDACs): Evolution, Specificity, Role in Transcriptional Complexes, and Pharmacological Actionability. Genes 2020, 11, 556. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhu, W.G. Targeting histone deacetylases for cancer therapy: From molecular mechanisms to clinical implications. Int. J. Biol. Sci. 2014, 10, 757–770. [Google Scholar] [CrossRef] [PubMed]
- Seto, E.; Yoshida, M. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6, a018713. [Google Scholar] [CrossRef]
- Falkenberg, K.J.; Johnstone, R.W. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat. Rev. Drug Discov. 2014, 13, 673–691. [Google Scholar] [CrossRef]
- Yoon, S.; Eom, G.H. HDAC and HDAC Inhibitor: From Cancer to Cardiovascular Diseases. Chonnam Med. J. 2016, 52, 1–11. [Google Scholar] [CrossRef]
- Brindisi, M.; Senger, J.; Cavella, C.; Grillo, A.; Chemi, G.; Gemma, S.; Cucinella, D.M.; Lamponi, S.; Sarno, F.; Iside, C.; et al. Novel spiroindoline HDAC inhibitors: Synthesis, molecular modelling and biological studies. Eur. J. Med. Chem. 2018, 157, 127–138. [Google Scholar] [CrossRef]
- Nian, H.; Bisson, W.H.; Dashwood, W.M.; Pinto, J.T.; Dashwood, R.H. Alpha-keto acid metabolites of organoselenium compounds inhibit histone deacetylase activity in human colon cancer cells. Carcinogenesis 2009, 30, 1416–1423. [Google Scholar] [CrossRef]
- Chakrabarti, A.; Oehme, I.; Witt, O.; Oliveira, G.; Sippl, W.; Romier, C.; Pierce, R.J.; Jung, M. HDAC8: A multifaceted target for therapeutic interventions. Trends Pharmacol. Sci. 2015, 36, 481–492. [Google Scholar] [CrossRef]
- Chakrabarti, A.; Melesina, J.; Kolbinger, F.R.; Oehme, I.; Senger, J.; Witt, O.; Sippl, W.; Jung, M. Targeting histone deacetylase 8 as a therapeutic approach to cancer and neurodegenerative diseases. Future Med. Chem. 2016, 8, 1609–1634. [Google Scholar] [CrossRef]
- Nalawansha, D.A.; Pflum, M.K. LSD1 Substrate Binding and Gene Expression Are Affected by HDAC1-Mediated Deacetylation. ACS Chem. Biol. 2017, 12, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Pan, H.; Montgomery, R.L.; Olson, E.N.; Schultz, R.M. Compensatory functions of histone deacetylase 1 (HDAC1) and HDAC2 regulate transcription and apoptosis during mouse oocyte development. Proc. Natl. Acad. Sci. USA 2012, 109, E481–E489. [Google Scholar] [CrossRef] [PubMed]
- Segré, C.V.; Chiocca, S. Regulating the regulators: The post-translational code of class I HDAC1 and HDAC2. J. Biomed. Biotechnol. 2011, 2011, 690848. [Google Scholar] [CrossRef] [PubMed]
- Bhaskara, S.; Chyla, B.J.; Amann, J.M.; Knutson, S.K.; Cortez, D.; Sun, Z.W.; Hiebert, S.W. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol. Cell 2008, 30, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Cho, H.; Yoo, J.; Kim, G.W.; Jeon, Y.H.; Lee, S.W.; Kwon, S.H. Pathological Role of HDAC8: Cancer and Beyond. Cells 2022, 11, 3161. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Kundu, S.; Singh, A.; Singh, S. Understanding the Role of Histone Deacetylase and their Inhibitors in Neurodegenerative Disorders: Current Targets and Future Perspective. Curr. Neuropharmacol. 2022, 20, 158–178. [Google Scholar] [CrossRef] [PubMed]
- McClure, J.J.; Li, X.; Chou, C.J. Advances and Challenges of HDAC Inhibitors in Cancer Therapeutics. Adv. Cancer Res. 2018, 138, 183–211. [Google Scholar] [CrossRef]
- Verdin, E.; Dequiedt, F.; Kasler, H. HDAC7 regulates apoptosis in developing thymocytes. Novartis Found. Symp. 2004, 259, 115–129; discussion 129–131, 163–169. [Google Scholar]
- Das, S.; Natarajan, R. HDAC9: An Inflammatory Link in Atherosclerosis. Circ. Res. 2020, 127, 824–826. [Google Scholar] [CrossRef]
- LoPresti, P. HDAC6 in Diseases of Cognition and of Neurons. Cells 2020, 10, 12. [Google Scholar] [CrossRef]
- Cheng, F.; Zheng, B.; Wang, J.; Zhao, G.; Yao, Z.; Niu, Z.; He, W. Histone deacetylase 10, a potential epigenetic target for therapy. Biosci. Rep. 2021, 41, BSR20210462. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Xie, C.; Chen, Q.; Zhuang, S. HDAC11, an emerging therapeutic target for metabolic disorders. Front. Endocrinol. 2022, 13, 989305. [Google Scholar] [CrossRef] [PubMed]
- Morigi, M.; Perico, L.; Benigni, A. Sirtuins in Renal Health and Disease. J. Am. Soc. Nephrol. 2018, 29, 1799–1809. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, D.; Feng, Y.; Li, B.; Cui, Y.; Chen, G.; Li, N. Association of sirtuins (SIRT1-7) with lung and intestinal diseases. Mol. Cell. Biochem. 2022, 477, 2539–2552. [Google Scholar] [CrossRef] [PubMed]
- Wątroba, M.; Dudek, I.; Skoda, M.; Stangret, A.; Rzodkiewicz, P.; Szukiewicz, D. Sirtuins, epigenetics and longevity. Ageing Res. Rev. 2017, 40, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Somoza, J.R.; Skene, R.J.; Katz, B.A.; Mol, C.; Ho, J.D.; Jennings, A.J.; Luong, C.; Arvai, A.; Buggy, J.J.; Chi, E.; et al. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure 2004, 12, 1325–1334. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Wang, S.; Zhou, N.; Cao, Z.; Zhang, Y. A proton-shuttle reaction mechanism for histone deacetylase 8 and the catalytic role of metal ions. J. Am. Chem. Soc. 2010, 132, 9471–9479. [Google Scholar] [CrossRef] [PubMed]
- Sauve, A.A.; Wolberger, C.; Schramm, V.L.; Boeke, J.D. The biochemistry of sirtuins. Annu. Rev. Biochem. 2006, 75, 435–465. [Google Scholar] [CrossRef] [PubMed]
- Michan, S.; Sinclair, D. Sirtuins in mammals: Insights into their biological function. Biochem. J. 2007, 404, 1–13. [Google Scholar] [CrossRef]
- Feldman, J.L.; Dittenhafer-Reed, K.E.; Denu, J.M. Sirtuin catalysis and regulation. J. Biol. Chem. 2012, 287, 42419–42427. [Google Scholar] [CrossRef]
- Ren, J.; Zhang, J.; Cai, H.; Li, Y.; Zhang, Y.; Zhang, X.; Zhao, D.; Li, Z.; Ma, H.; Wang, J.; et al. HDAC as a therapeutic target for treatment of endometrial cancers. Curr. Pharm. Des. 2014, 20, 1847–1856. [Google Scholar] [CrossRef] [PubMed]
- Witt, O.; Deubzer, H.E.; Milde, T.; Oehme, I. HDAC family: What are the cancer relevant targets? Cancer Lett. 2009, 277, 8–21. [Google Scholar] [CrossRef] [PubMed]
- de Ruijter, A.J.; van Gennip, A.H.; Caron, H.N.; Kemp, S.; van Kuilenburg, A.B. Histone deacetylases (HDACs): Characterization of the classical HDAC family. Biochem. J. 2003, 370, 737–749. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.J.; Seto, E. The Rpd3/Hda1 family of lysine deacetylases: From bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 2008, 9, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Pflum, M.K.; Tong, J.K.; Lane, W.S.; Schreiber, S.L. Histone deacetylase 1 phosphorylation promotes enzymatic activity and complex formation. J. Biol. Chem. 2001, 276, 47733–47741. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, R.; Banerjee, S.; Amin, S.A.; Adhikari, N.; Jha, T. Histone deacetylase 3 (HDAC3) inhibitors as anticancer agents: A review. Eur. J. Med. Chem. 2020, 192, 112171. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.M.; Tsai, S.C.; Wen, Y.D.; Fejer, G.; Seto, E. Functional domains of histone deacetylase-3. J. Biol. Chem. 2002, 277, 9447–9454. [Google Scholar] [CrossRef] [PubMed]
- Minucci, S.; Pelicci, P.G. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat. Rev. Cancer 2006, 6, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Bondarev, A.D.; Attwood, M.M.; Jonsson, J.; Chubarev, V.N.; Tarasov, V.V.; Schiöth, H.B. Recent developments of HDAC inhibitors: Emerging indications and novel molecules. Br. J. Clin. Pharmacol. 2021, 87, 4577–4597. [Google Scholar] [CrossRef]
- De Souza, C.; Chatterji, B.P. HDAC Inhibitors as Novel Anti-Cancer Therapeutics. Recent. Pat. Anticancer Drug Discov. 2015, 10, 145–162. [Google Scholar] [CrossRef]
- Li, J.; Lu, L.; Liu, L.; Ren, X.; Chen, J.; Yin, X.; Xiao, Y.; Wei, G.; Huang, H.; Wei, W.; et al. HDAC1/2/3 are major histone desuccinylases critical for promoter desuccinylation. Cell Discov. 2023, 9, 85. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.A.; Adhikari, N.; Jha, T. Structure-activity relationships of HDAC8 inhibitors: Non-hydroxamates as anticancer agents. Pharmacol. Res. 2018, 131, 128–142. [Google Scholar] [CrossRef] [PubMed]
- Watson, P.J.; Fairall, L.; Santos, G.M.; Schwabe, J.W. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature 2012, 481, 335–340. [Google Scholar] [CrossRef]
- Millard, C.J.; Watson, P.J.; Celardo, I.; Gordiyenko, Y.; Cowley, S.M.; Robinson, C.V.; Fairall, L.; Schwabe, J.W. Class I HDACs share a common mechanism of regulation by inositol phosphates. Mol. Cell 2013, 51, 57–67. [Google Scholar] [CrossRef]
- Lauffer, B.E.; Mintzer, R.; Fong, R.; Mukund, S.; Tam, C.; Zilberleyb, I.; Flicke, B.; Ritscher, A.; Fedorowicz, G.; Vallero, R.; et al. Histone deacetylase (HDAC) inhibitor kinetic rate constants correlate with cellular histone acetylation but not transcription and cell viability. J. Biol. Chem. 2013, 288, 26926–26943. [Google Scholar] [CrossRef] [PubMed]
- Bantscheff, M.; Hopf, C.; Savitski, M.M.; Dittmann, A.; Grandi, P.; Michon, A.M.; Schlegl, J.; Abraham, Y.; Becher, I.; Bergamini, G.; et al. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat. Biotechnol. 2011, 29, 255–265. [Google Scholar] [CrossRef]
- Hsu, K.C.; Liu, C.Y.; Lin, T.E.; Hsieh, J.H.; Sung, T.Y.; Tseng, H.J.; Yang, J.M.; Huang, W.J. Novel Class IIa-Selective Histone Deacetylase Inhibitors Discovered Using an in Silico Virtual Screening Approach. Sci. Rep. 2017, 7, 3228. [Google Scholar] [CrossRef]
- Liu, L.; Dong, L.; Bourguet, E.; Fairlie, D.P. Targeting Class IIa HDACs: Insights from Phenotypes and Inhibitors. Curr. Med. Chem. 2021, 28, 8628–8672. [Google Scholar] [CrossRef]
- Wright, L.H.; Menick, D.R. A class of their own: Exploring the nondeacetylase roles of class IIa HDACs in cardiovascular disease. Am. J. Physiol. Heart Circ. Physiol. 2016, 311, H199–H206. [Google Scholar] [CrossRef]
- Hess, L.; Moos, V.; Lauber, A.A.; Reiter, W.; Schuster, M.; Hartl, N.; Lackner, D.; Boenke, T.; Koren, A.; Guzzardo, P.M.; et al. A toolbox for class I HDACs reveals isoform specific roles in gene regulation and protein acetylation. PLoS Genet. 2022, 18, e1010376. [Google Scholar] [CrossRef]
- Park, S.Y.; Kim, G.S.; Hwang, H.J.; Nam, T.H.; Park, H.S.; Song, J.; Jang, T.H.; Lee, Y.C.; Kim, J.S. Structural basis of the specific interaction of SMRT corepressor with histone deacetylase 4. Nucleic Acids Res. 2018, 46, 11776–11788. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Xiao, J.; Wang, Y.; Song, X.; Huang, L.; Ren, Z.; Kitazato, K. Posttranslational modification and beyond: Interplay between histone deacetylase 6 and heat-shock protein 90. Mol. Med. 2021, 27, 110. [Google Scholar] [CrossRef] [PubMed]
- Yue, K.; Qin, M.; Huang, C.; James Chou, C.; Jiang, Y.; Li, X. Comparison of three zinc binding groups for HDAC inhibitors—A potency, selectivity and enzymatic kinetics study. Bioorg Med. Chem. Lett. 2022, 70, 128797. [Google Scholar] [CrossRef] [PubMed]
- Bertos, N.R.; Gilquin, B.; Chan, G.K.; Yen, T.J.; Khochbin, S.; Yang, X.J. Role of the tetradecapeptide repeat domain of human histone deacetylase 6 in cytoplasmic retention. J. Biol. Chem. 2004, 279, 48246–48254. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Zhang, F.; Maguire, A.; Byrne, T.; Weiner-Gorzel, K.; Bridgett, S.; O’Toole, S.; O’Leary, J.; Beggan, C.; Fitzpatrick, P.; et al. HDAC6 Degradation Inhibits the Growth of High-Grade Serous Ovarian Cancer Cells. Cancers 2020, 12, 3734. [Google Scholar] [CrossRef] [PubMed]
- Losson, H.; Schnekenburger, M.; Dicato, M.; Diederich, M. HDAC6-an Emerging Target Against Chronic Myeloid Leukemia? Cancers 2020, 12, 318. [Google Scholar] [CrossRef] [PubMed]
- English, K.; Barton, M.C. HDAC6: A Key Link Between Mitochondria and Development of Peripheral Neuropathy. Front. Mol. Neurosci. 2021, 14, 684714. [Google Scholar] [CrossRef]
- Herp, D.; Ridinger, J.; Robaa, D.; Shinsky, S.A.; Schmidtkunz, K.; Yesiloglu, T.Z.; Bayer, T.; Steimbach, R.R.; Herbst-Gervasoni, C.J.; Merz, A.; et al. First Fluorescent Acetylspermidine Deacetylation Assay for HDAC10 Identifies Selective Inhibitors with Cellular Target Engagement. ChemBioChem 2022, 23, e202200180. [Google Scholar] [CrossRef]
- Bottomley, M.J.; Lo Surdo, P.; Di Giovine, P.; Cirillo, A.; Scarpelli, R.; Ferrigno, F.; Jones, P.; Neddermann, P.; De Francesco, R.; Steinkühler, C.; et al. Structural and functional analysis of the human HDAC4 catalytic domain reveals a regulatory structural zinc-binding domain. J. Biol. Chem. 2008, 283, 26694–26704. [Google Scholar] [CrossRef]
- Hai, Y.; Christianson, D.W. Histone deacetylase 6 structure and molecular basis of catalysis and inhibition. Nat. Chem. Biol. 2016, 12, 741–747. [Google Scholar] [CrossRef]
- Schuetz, A.; Min, J.; Allali-Hassani, A.; Schapira, M.; Shuen, M.; Loppnau, P.; Mazitschek, R.; Kwiatkowski, N.P.; Lewis, T.A.; Maglathin, R.L.; et al. Human HDAC7 harbors a class IIa histone deacetylase-specific zinc binding motif and cryptic deacetylase activity. J. Biol. Chem. 2008, 283, 11355–11363. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.S.; Wu, F.; Jin, Y.M.; Chang, W.Q.; Xu, T.M. HDAC11: A rising star in epigenetics. Biomed. Pharmacother. 2020, 131, 110607. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tong, X.; Hu, W.; Chen, D. HDAC11: A novel target for improved cancer therapy. Biomed. Pharmacother. 2023, 166, 115418. [Google Scholar] [CrossRef] [PubMed]
- Yanginlar, C.; Logie, C. HDAC11 is a regulator of diverse immune functions. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Blander, G.; Guarente, L. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 2004, 73, 417–435. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.J.; Zhang, T.N.; Chen, H.H.; Yu, X.F.; Lv, J.L.; Liu, Y.Y.; Liu, Y.S.; Zheng, G.; Zhao, J.Q.; Wei, Y.F.; et al. The sirtuin family in health and disease. Signal Transduct. Target. Ther. 2022, 7, 402. [Google Scholar] [CrossRef] [PubMed]
- Villalba, J.M.; Alcaín, F.J. Sirtuin activators and inhibitors. Biofactors 2012, 38, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Gasparrini, M.; Mazzola, F.; Cuccioloni, M.; Sorci, L.; Audrito, V.; Zamporlini, F.; Fortunato, C.; Amici, A.; Cianci, M.; Deaglio, S.; et al. Molecular insights into the interaction between human nicotinamide phosphoribosyltransferase and Toll-like receptor 4. J. Biol. Chem. 2022, 298, 101669. [Google Scholar] [CrossRef]
- Simó-Mirabet, P.; Bermejo-Nogales, A.; Calduch-Giner, J.A.; Pérez-Sánchez, J. Tissue-specific gene expression and fasting regulation of sirtuin family in gilthead sea bream (Sparus aurata). J. Comp. Physiol. B 2017, 187, 153–163. [Google Scholar] [CrossRef]
- Chen, D.; Guarente, L. SIR2: A potential target for calorie restriction mimetics. Trends Mol. Med. 2007, 13, 64–71. [Google Scholar] [CrossRef]
- Arellano-Ballestero, H.; Sabry, M.; Lowdell, M.W. A Killer Disarmed: Natural Killer Cell Impairment in Myelodysplastic Syndrome. Cells 2023, 12, 633. [Google Scholar] [CrossRef] [PubMed]
- Lucatelli, P.; De Rubeis, G.; Trobiani, C.; Ungania, S.; Rocco, B.; De Gyurgyokai, S.Z.; Masi, M.; Pecorella, I.; Cappelli, F.; Lai, Q.; et al. In Vivo Comparison of Micro-Balloon Interventions (MBI) Advantage: A Retrospective Cohort Study of DEB-TACE Versus b-TACE and of SIRT Versus b-SIRT. Cardiovasc. Intervent Radiol. 2022, 45, 306–314. [Google Scholar] [CrossRef]
- Zhang, J.; Xiang, H.; Liu, J.; Chen, Y.; He, R.R.; Liu, B. Mitochondrial Sirtuin 3: New emerging biological function and therapeutic target. Theranostics 2020, 10, 8315–8342. [Google Scholar] [CrossRef] [PubMed]
- Di Emidio, G.; Falone, S.; Artini, P.G.; Amicarelli, F.; D’Alessandro, A.M.; Tatone, C. Mitochondrial Sirtuins in Reproduction. Antioxidants 2021, 10, 1047. [Google Scholar] [CrossRef]
- Kida, Y.; Goligorsky, M.S. Sirtuins, Cell Senescence, and Vascular Aging. Can. J. Cardiol. 2016, 32, 634–641. [Google Scholar] [CrossRef] [PubMed]
- Beegum, F.; Anuranjana, P.V.; George, K.T.; KP, D.; Begum, F.; Krishnadas, N.; Shenoy, R.R. Sirtuins as therapeutic targets for improving delayed wound healing in diabetes. J. Drug Target. 2022, 30, 911–926. [Google Scholar] [CrossRef]
- Zhao, X.; Allison, D.; Condon, B.; Zhang, F.; Gheyi, T.; Zhang, A.; Ashok, S.; Russell, M.; MacEwan, I.; Qian, Y.; et al. The 2.5 Å crystal structure of the SIRT1 catalytic domain bound to nicotinamide adenine dinucleotide (NAD+) and an indole (EX527 analogue) reveals a novel mechanism of histone deacetylase inhibition. J. Med. Chem. 2013, 56, 963–969. [Google Scholar] [CrossRef]
- Moniot, S.; Schutkowski, M.; Steegborn, C. Crystal structure analysis of human Sirt2 and its ADP-ribose complex. J. Struct. Biol. 2013, 182, 136–143. [Google Scholar] [CrossRef]
- Nguyen, G.T.; Schaefer, S.; Gertz, M.; Weyand, M.; Steegborn, C. Structures of human sirtuin 3 complexes with ADP-ribose and with carba-NAD+ and SRT1720: Binding details and inhibition mechanism. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 1423–1432. [Google Scholar] [CrossRef]
- Li, G.-B.; Deng, J. Crystal Structure of Human SIRT5 in Complex with Diazidine Inhibitor 9. 2023. [Google Scholar] [CrossRef]
- Rajabi, N.; Auth, M.; Troelsen, K.R.; Pannek, M.; Bhatt, D.P.; Fontenas, M.; Hirschey, M.D.; Steegborn, C.; Madsen, A.S.; Olsen, C.A. Mechanism-Based Inhibitors of the Human Sirtuin 5 Deacylase: Structure-Activity Relationship, Biostructural, and Kinetic Insight. Angew. Chem. Int. Ed. Engl. 2017, 56, 14836–14841. [Google Scholar] [CrossRef]
- West, A.C.; Johnstone, R.W. New and emerging HDAC inhibitors for cancer treatment. J. Clin. Investig. 2014, 124, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.; Tekwani, B.L. Histone Deacetylases Inhibitors in Neurodegenerative Diseases, Neuroprotection and Neuronal Differentiation. Front. Pharmacol. 2020, 11, 537. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Ge, J.; Li, H. Lysine acetyltransferases and lysine deacetylases as targets for cardiovascular disease. Nat. Rev. Cardiol. 2020, 17, 96–115. [Google Scholar] [CrossRef]
- Kulthinee, S.; Yano, N.; Zhuang, S.; Wang, L.; Zhao, T.C. Critical Functions of Histone Deacetylases (HDACs) in Modulating Inflammation Associated with Cardiovascular Diseases. Pathophysiology 2022, 29, 471–485. [Google Scholar] [CrossRef] [PubMed]
- Carafa, V.; Nebbioso, A.; Altucci, L. Sirtuins and disease: The road ahead. Front. Pharmacol. 2012, 3, 4. [Google Scholar] [CrossRef]
- Zeng, L.S.; Yang, X.Z.; Wen, Y.F.; Mail, S.J.; Wang, M.H.; Zhang, M.Y.; Zheng, X.F.; Wang, H.Y. Overexpressed HDAC4 is associated with poor survival and promotes tumor progression in esophageal carcinoma. Aging 2016, 8, 1236–1249. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Huang, Y.; Wang, Z.; Wang, H.T.; Duan, B.; Ye, D.; Wang, C.; Jing, R.; Leng, Y.; Xi, J.; et al. HDAC10 promotes lung cancer proliferation via AKT phosphorylation. Oncotarget 2016, 7, 59388–59401. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, X.; Zhu, S.; Dejene, E.A.; Peng, W.; Sepulveda, A.; Seto, E. HDAC10 Regulates Cancer Stem-Like Cell Properties in KRAS-Driven Lung Adenocarcinoma. Cancer Res. 2020, 80, 3265–3278. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Han, S.; Cheng, W.; Miao, R.; Li, S.; Tian, X.; Kan, Q. Design, Synthesis, and Biological Evaluation of 2-Anilino-4-Triazolpyrimidine Derivatives as CDK4/HDACs Inhibitors. Drug Des. Dev. Ther. 2022, 16, 1083–1097. [Google Scholar] [CrossRef]
- Morse, J.S.; Sheng, Y.J.; Hampton, J.T.; Sylvain, L.D.; Das, S.; Alugubelli, Y.R.; Chen, P.C.; Yang, K.S.; Xu, S.; Fierke, C.A.; et al. Phage-assisted, active site-directed ligand evolution of a potent and selective histone deacetylase 8 inhibitor. Protein Sci. 2022, 31, e4512. [Google Scholar] [CrossRef]
- Bertrand, P. Inside HDAC with HDAC inhibitors. Eur. J. Med. Chem. 2010, 45, 2095–2116. [Google Scholar] [CrossRef] [PubMed]
- Murayama, A.; Ohmori, K.; Fujimura, A.; Minami, H.; Yasuzawa-Tanaka, K.; Kuroda, T.; Oie, S.; Daitoku, H.; Okuwaki, M.; Nagata, K.; et al. Epigenetic control of rDNA loci in response to intracellular energy status. Cell 2008, 133, 627–639. [Google Scholar] [CrossRef] [PubMed]
- Peserico, A.; Chiacchiera, F.; Grossi, V.; Matrone, A.; Latorre, D.; Simonatto, M.; Fusella, A.; Ryall, J.G.; Finley, L.W.; Haigis, M.C.; et al. A novel AMPK-dependent FoxO3A-SIRT3 intramitochondrial complex sensing glucose levels. Cell. Mol. Life Sci. 2013, 70, 2015–2029. [Google Scholar] [CrossRef] [PubMed]
- Mahlknecht, U.; Ho, A.D.; Letzel, S.; Voelter-Mahlknecht, S. Assignment of the NAD-dependent deacetylase sirtuin 5 gene (SIRT5) to human chromosome band 6p23 by in situ hybridization. Cytogenet. Genome Res. 2006, 112, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Voelter-Mahlknecht, S.; Letzel, S.; Mahlknecht, U. Fluorescence in situ hybridization and chromosomal organization of the human Sirtuin 7 gene. Int. J. Oncol. 2006, 28, 899–908. [Google Scholar] [CrossRef] [PubMed]
- Mathias, R.A.; Guise, A.J.; Cristea, I.M. Post-translational modifications regulate class IIa histone deacetylase (HDAC) function in health and disease. Mol. Cell. Proteom. 2015, 14, 456–470. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, N.; Seto, E. Regulation of histone deacetylase activities. J. Cell. Biochem. 2004, 93, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Fischle, W.; Dequiedt, F.; Hendzel, M.J.; Guenther, M.G.; Lazar, M.A.; Voelter, W.; Verdin, E. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell 2002, 9, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Qin, G.; Zhao, T.C. HDAC4: Mechanism of regulation and biological functions. Epigenomics 2014, 6, 139–150. [Google Scholar] [CrossRef]
- Roy, D.M.; Walsh, L.A.; Chan, T.A. Driver mutations of cancer epigenomes. Protein Cell 2014, 5, 265–296. [Google Scholar] [CrossRef]
- Berger, M.F.; Lawrence, M.S.; Demichelis, F.; Drier, Y.; Cibulskis, K.; Sivachenko, A.Y.; Sboner, A.; Esgueva, R.; Pflueger, D.; Sougnez, C.; et al. The genomic complexity of primary human prostate cancer. Nature 2011, 470, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Ropero, S.; Fraga, M.F.; Ballestar, E.; Hamelin, R.; Yamamoto, H.; Boix-Chornet, M.; Caballero, R.; Alaminos, M.; Setien, F.; Paz, M.F.; et al. A truncating mutation of HDAC2 in human cancers confers resistance to histone deacetylase inhibition. Nat. Genet. 2006, 38, 566–569. [Google Scholar] [CrossRef] [PubMed]
- Sjöblom, T.; Jones, S.; Wood, L.D.; Parsons, D.W.; Lin, J.; Barber, T.D.; Mandelker, D.; Leary, R.J.; Ptak, J.; Silliman, N.; et al. The consensus coding sequences of human breast and colorectal cancers. Science 2006, 314, 268–274. [Google Scholar] [CrossRef]
- Ropero, S.; Ballestar, E.; Alaminos, M.; Arango, D.; Schwartz, S.; Esteller, M. Transforming pathways unleashed by a HDAC2 mutation in human cancer. Oncogene 2008, 27, 4008–4012. [Google Scholar] [CrossRef]
- Jones, S.; Wang, T.L.; Shih, I.M.; Mao, T.L.; Nakayama, K.; Roden, R.; Glas, R.; Slamon, D.; Diaz, L.A.; Vogelstein, B.; et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 2010, 330, 228–231. [Google Scholar] [CrossRef]
- Wiegand, K.C.; Shah, S.P.; Al-Agha, O.M.; Zhao, Y.; Tse, K.; Zeng, T.; Senz, J.; McConechy, M.K.; Anglesio, M.S.; Kalloger, S.E.; et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med. 2010, 363, 1532–1543. [Google Scholar] [CrossRef] [PubMed]
- Weichert, W.; Denkert, C.; Noske, A.; Darb-Esfahani, S.; Dietel, M.; Kalloger, S.E.; Huntsman, D.G.; Köbel, M. Expression of class I histone deacetylases indicates poor prognosis in endometrioid subtypes of ovarian and endometrial carcinomas. Neoplasia 2008, 10, 1021–1027. [Google Scholar] [CrossRef]
- Fukumoto, T.; Park, P.H.; Wu, S.; Fatkhutdinov, N.; Karakashev, S.; Nacarelli, T.; Kossenkov, A.V.; Speicher, D.W.; Jean, S.; Zhang, L.; et al. Repurposing Pan-HDAC Inhibitors for ARID1A-Mutated Ovarian Cancer. Cell Rep. 2018, 22, 3393–3400. [Google Scholar] [CrossRef]
- Kobayashi, T.; Nakazono, K.; Tokuda, M.; Mashima, Y.; Dynlacht, B.D.; Itoh, H. HDAC2 promotes loss of primary cilia in pancreatic ductal adenocarcinoma. EMBO Rep. 2017, 18, 334–343. [Google Scholar] [CrossRef]
- King, J.; Patel, M.; Chandrasekaran, S. Metabolism, HDACs, and HDAC Inhibitors: A Systems Biology Perspective. Metabolites 2021, 11, 792. [Google Scholar] [CrossRef]
- Annibaldi, A.; Widmann, C. Glucose metabolism in cancer cells. Curr. Opin. Clin. Nutr. Metab. Care 2010, 13, 466–470. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Jin, X.; Yan, Y.; Shao, Y.; Pan, Y.; Roberts, L.R.; Zhang, J.; Huang, H.; Jiang, J. Inhibiting histone deacetylases suppresses glucose metabolism and hepatocellular carcinoma growth by restoring FBP1 expression. Sci. Rep. 2017, 7, 43864. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Sun, Z. Mechanism of Action for HDAC Inhibitors-Insights from Omics Approaches. Int. J. Mol. Sci. 2019, 20, 1616. [Google Scholar] [CrossRef]
- Zhu, M.; Han, Y.; Gu, T.; Wang, R.; Si, X.; Kong, D.; Zhao, P.; Wang, X.; Li, J.; Zhai, X.; et al. Class I HDAC inhibitors enhance antitumor efficacy and persistence of CAR-T cells by activation of the Wnt pathway. Cell Rep. 2024, 43, 114065. [Google Scholar] [CrossRef] [PubMed]
- Amoêdo, N.D.; Rodrigues, M.F.; Pezzuto, P.; Galina, A.; da Costa, R.M.; de Almeida, F.C.; El-Bacha, T.; Rumjanek, F.D. Energy metabolism in H460 lung cancer cells: Effects of histone deacetylase inhibitors. PLoS ONE 2011, 6, e22264. [Google Scholar] [CrossRef] [PubMed]
- Fadaka, A.; Ajiboye, B.; Ojo, O.; Adewale, O.; Olayide, I.; Emuowhochere, R. Biology of glucose metabolization in cancer cells. J. Oncol. Sci. 2017, 3, 45–51. [Google Scholar] [CrossRef]
- Furumai, R.; Komatsu, Y.; Nishino, N.; Khochbin, S.; Yoshida, M.; Horinouchi, S. Potent histone deacetylase inhibitors built from trichostatin A and cyclic tetrapeptide antibiotics including trapoxin. Proc. Natl. Acad. Sci. USA 2001, 98, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Kijima, M.; Yoshida, M.; Sugita, K.; Horinouchi, S.; Beppu, T. Trapoxin, an antitumor cyclic tetrapeptide, is an irreversible inhibitor of mammalian histone deacetylase. J. Biol. Chem. 1993, 268, 22429–22435. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, P.; Williams, D.E.; Ho, E.; Dashwood, R.H. Metabolism as a key to histone deacetylase inhibition. Crit. Rev. Biochem. Mol. Biol. 2011, 46, 181–199. [Google Scholar] [CrossRef]
- Desai, D.; Salli, U.; Vrana, K.E.; Amin, S. SelSA, selenium analogs of SAHA as potent histone deacetylase inhibitors. Bioorg. Med. Chem. Lett. 2010, 20, 2044–2047. [Google Scholar] [CrossRef]
- Sanaei, M.; Kavoosi, F. Histone Deacetylases and Histone Deacetylase Inhibitors: Molecular Mechanisms of Action in Various Cancers. Adv. Biomed. Res. 2019, 8, 63. [Google Scholar] [CrossRef] [PubMed]
- Ceccacci, E.; Minucci, S. Inhibition of histone deacetylases in cancer therapy: Lessons from leukaemia. Br. J. Cancer 2016, 114, 605–611. [Google Scholar] [CrossRef] [PubMed]
- Mottamal, M.; Zheng, S.; Huang, T.L.; Wang, G. Histone deacetylase inhibitors in clinical studies as templates for new anticancer agents. Molecules 2015, 20, 3898–3941. [Google Scholar] [CrossRef] [PubMed]
- Gilardini Montani, M.S.; Granato, M.; Santoni, C.; Del Porto, P.; Merendino, N.; D’Orazi, G.; Faggioni, A.; Cirone, M. Histone deacetylase inhibitors VPA and TSA induce apoptosis and autophagy in pancreatic cancer cells. Cell. Oncol. 2017, 40, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Hao, C.; Wang, L.; Liu, P.; Zhao, L.; Zhu, C.; Tian, X. Inhibition of leukemic cells by valproic acid, an HDAC inhibitor, in xenograft tumors. OncoTargets Ther. 2013, 6, 733–740. [Google Scholar] [CrossRef]
- Johnstone, R.W.; Ruefli, A.A.; Lowe, S.W. Apoptosis: A link between cancer genetics and chemotherapy. Cell 2002, 108, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, M.; Furumai, R.; Nishiyama, M.; Komatsu, Y.; Nishino, N.; Horinouchi, S. Histone deacetylase as a new target for cancer chemotherapy. Cancer Chemother. Pharmacol. 2001, 48 (Suppl. S1), S20–S26. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, X.; Zhang, E.; Yang, L.; Yuan, H.; Tu, W.; Zhang, H.; Yin, Z.; Shen, W.; Chen, X.; et al. An epigenetic bioactive composite scaffold with well-aligned nanofibers for functional tendon tissue engineering. Acta Biomater. 2018, 66, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Kelly, W.K.; O’Connor, O.A.; Krug, L.M.; Chiao, J.H.; Heaney, M.; Curley, T.; MacGregore-Cortelli, B.; Tong, W.; Secrist, J.P.; Schwartz, L.; et al. Phase I study of an oral histone deacetylase inhibitor, suberoylanilide hydroxamic acid, in patients with advanced cancer. J. Clin. Oncol. 2005, 23, 3923–3931. [Google Scholar] [CrossRef]
- O’Connor, O.A.; Heaney, M.L.; Schwartz, L.; Richardson, S.; Willim, R.; MacGregor-Cortelli, B.; Curly, T.; Moskowitz, C.; Portlock, C.; Horwitz, S.; et al. Clinical experience with intravenous and oral formulations of the novel histone deacetylase inhibitor suberoylanilide hydroxamic acid in patients with advanced hematologic malignancies. J. Clin. Oncol. 2006, 24, 166–173. [Google Scholar] [CrossRef]
- Ho, T.C.S.; Chan, A.H.Y.; Ganesan, A. Thirty Years of HDAC Inhibitors: 2020 Insight and Hindsight. J. Med. Chem. 2020, 63, 12460–12484. [Google Scholar] [CrossRef]
- Finnin, M.S.; Donigian, J.R.; Cohen, A.; Richon, V.M.; Rifkind, R.A.; Marks, P.A.; Breslow, R.; Pavletich, N.P. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 1999, 401, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Miller, T.A.; Witter, D.J.; Belvedere, S. Histone deacetylase inhibitors. J. Med. Chem. 2003, 46, 5097–5116. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Lu, Z.; Cao, Z.; Zhang, Y. Zinc chelation with hydroxamate in histone deacetylases modulated by water access to the linker binding channel. J. Am. Chem. Soc. 2011, 133, 6110–6113. [Google Scholar] [CrossRef] [PubMed]
- Rajak, H.; Singh, A.; Raghuwanshi, K.; Kumar, R.; Dewangan, P.K.; Veerasamy, R.; Sharma, P.C.; Dixit, A.; Mishra, P. A structural insight into hydroxamic acid based histone deacetylase inhibitors for the presence of anticancer activity. Curr. Med. Chem. 2014, 21, 2642–2664. [Google Scholar] [CrossRef] [PubMed]
- Micelli, C.; Rastelli, G. Histone deacetylases: Structural determinants of inhibitor selectivity. Drug Discov. Today 2015, 20, 718–735. [Google Scholar] [CrossRef]
- Heers, H.; Stanislaw, J.; Harrelson, J.; Lee, M.W. Valproic acid as an adjunctive therapeutic agent for the treatment of breast cancer. Eur. J. Pharmacol. 2018, 835, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Paredes, M.; Esteller, M. Cancer epigenetics reaches mainstream oncology. Nat. Med. 2011, 17, 330–339. [Google Scholar] [CrossRef]
- Li, Y.; Seto, E. HDACs and HDAC Inhibitors in Cancer Development and Therapy. Cold Spring Harb. Perspect. Med. 2016, 6, a026831. [Google Scholar] [CrossRef]
- Manal, M.; Chandrasekar, M.J.; Gomathi Priya, J.; Nanjan, M.J. Inhibitors of histone deacetylase as antitumor agents: A critical review. Bioorg. Chem. 2016, 67, 18–42. [Google Scholar] [CrossRef]
- Zhang, L.; Han, Y.; Jiang, Q.; Wang, C.; Chen, X.; Li, X.; Xu, F.; Jiang, Y.; Wang, Q.; Xu, W. Trend of histone deacetylase inhibitors in cancer therapy: Isoform selectivity or multitargeted strategy. Med. Res. Rev. 2015, 35, 63–84. [Google Scholar] [CrossRef]
- Giannini, G.; Cabri, W.; Fattorusso, C.; Rodriquez, M. Histone deacetylase inhibitors in the treatment of cancer: Overview and perspectives. Future Med. Chem. 2012, 4, 1439–1460. [Google Scholar] [CrossRef] [PubMed]
- Mann, B.S.; Johnson, J.R.; Cohen, M.H.; Justice, R.; Pazdur, R. FDA approval summary: Vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist 2007, 12, 1247–1252. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.Y.; Ding, W.; Li, T.Q.; Zhang, Y.X.; Zhao, S.C. Histone Deacetylase (HDAC) Inhibitor, Suberoylanilide Hydroxamic Acid (SAHA), Induces Apoptosis in Prostate Cancer Cell Lines via the Akt/FOXO3a Signaling Pathway. Med. Sci. Monit. 2017, 23, 5793–5802. [Google Scholar] [CrossRef] [PubMed]
- Mrakovcic, M.; Kleinheinz, J.; Fröhlich, L.F. Histone Deacetylase Inhibitor-Induced Autophagy in Tumor Cells: Implications for p53. Int. J. Mol. Sci. 2017, 18, 1883. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Zhao, N.; Hu, Y.; Jiang, C.S.; Zhang, H. The Development Process: From SAHA to Hydroxamate HDAC Inhibitors with Branched CAP Region and Linear Linker. Chem. Biodivers. 2020, 17, e1900427. [Google Scholar] [CrossRef] [PubMed]
- Brightman, A.O.; Wang, J.; Miu, R.K.; Sun, I.L.; Barr, R.; Crane, F.L.; Morré, D.J. A growth factor- and hormone-stimulated NADH oxidase from rat liver plasma membrane. Biochim. Biophys. Acta 1992, 1105, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.; Wahan, S.K.; Vishakha, S.; Kurmi, B.D.; Gupta, G.D.; Rajak, H.; Asati, V. Recent Progress in Histone Deacetylase (HDAC) 1 Inhibitors as Anticancer Agent. Curr. Cancer Drug Targets 2022, 23, 47–70. [Google Scholar] [CrossRef] [PubMed]
- Bass, A.K.A.; El-Zoghbi, M.S.; Nageeb, E.M.; Mohamed, M.F.A.; Badr, M.; Abuo-Rahma, G.E.A. Comprehensive review for anticancer hybridized multitargeting HDAC inhibitors. Eur. J. Med. Chem. 2021, 209, 112904. [Google Scholar] [CrossRef]
- Bressi, J.C.; Jennings, A.J.; Skene, R.; Wu, Y.; Melkus, R.; De Jong, R.; O’Connell, S.; Grimshaw, C.E.; Navre, M.; Gangloff, A.R. Exploration of the HDAC2 foot pocket: Synthesis and SAR of substituted N-(2-aminophenyl)benzamides. Bioorg. Med. Chem. Lett. 2010, 20, 3142–3145. [Google Scholar] [CrossRef]
- Dong, M.; Ning, Z.Q.; Xing, P.Y.; Xu, J.L.; Cao, H.X.; Dou, G.F.; Meng, Z.Y.; Shi, Y.K.; Lu, X.P.; Feng, F.Y. Phase I study of chidamide (CS055/HBI-8000), a new histone deacetylase inhibitor, in patients with advanced solid tumors and lymphomas. Cancer Chemother. Pharmacol. 2012, 69, 1413–1422. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, T.; Geng, C.; Zhang, Y.; Zhang, J.; Ning, Z.; Jiang, Z. Exploratory clinical study of chidamide, an oral subtype-selective histone deacetylase inhibitor, in combination with exemestane in hormone receptor-positive advanced breast cancer. Chin. J. Cancer Res. 2018, 30, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Buckton, L.K.; Rahimi, M.N.; McAlpine, S.R. Cyclic Peptides as Drugs for Intracellular Targets: The Next Frontier in Peptide Therapeutic Development. Chemistry 2021, 27, 1487–1513. [Google Scholar] [CrossRef] [PubMed]
- Ramadhani, D.; Maharani, R.; Gazzali, A.M.; Muchtaridi, M. Cyclic Peptides for the Treatment of Cancers: A Review. Molecules 2022, 27, 4428. [Google Scholar] [CrossRef] [PubMed]
- Pojani, E.; Barlocco, D. Romidepsin (FK228), A Histone Deacetylase Inhibitor and its Analogues in Cancer Chemotherapy. Curr. Med. Chem. 2021, 28, 1290–1303. [Google Scholar] [CrossRef] [PubMed]
- Ni, M.; Esposito, E.; Raj, V.P.; Muzi, L.; Zunino, F.; Zuco, V.; Cominetti, D.; Penco, S.; Dal Pozzo, A. New macrocyclic analogs of the natural histone deacetylase inhibitor FK228; design, synthesis and preliminary biological evaluation. Bioorg. Med. Chem. 2015, 23, 6785–6793. [Google Scholar] [CrossRef] [PubMed]
- Furumai, R.; Matsuyama, A.; Kobashi, N.; Lee, K.H.; Nishiyama, M.; Nakajima, H.; Tanaka, A.; Komatsu, Y.; Nishino, N.; Yoshida, M.; et al. FK228 (depsipeptide) as a natural prodrug that inhibits class I histone deacetylases. Cancer Res. 2002, 62, 4916–4921. [Google Scholar] [PubMed]
- Eckel, R.H.; Kahn, R.; Robertson, R.M.; Rizza, R.A. Preventing cardiovascular disease and diabetes: A call to action from the American Diabetes Association and the American Heart Association. Circulation 2006, 113, 2943–2946. [Google Scholar] [CrossRef] [PubMed]
- Sacks, F.M.; Lichtenstein, A.; Van Horn, L.; Harris, W.; Kris-Etherton, P.; Winston, M. Soy protein, isoflavones, and cardiovascular health: A summary of a statement for professionals from the american heart association nutrition committee. Arterioscler. Thromb. Vasc. Biol. 2006, 26, 1689–1692. [Google Scholar] [CrossRef]
- Wanczyk, M.; Roszczenko, K.; Marcinkiewicz, K.; Bojarczuk, K.; Kowara, M.; Winiarska, M. HDACi--going through the mechanisms. Front. Biosci. 2011, 16, 340–359. [Google Scholar] [CrossRef]
- Yang, R.; Lu, G.; Lv, Z.; Jia, L.; Cui, J. Different treatment regimens in breast cancer visceral crisis: A retrospective cohort study. Front. Oncol. 2022, 12, 1048781. [Google Scholar] [CrossRef]
- Tasneem, S.; Alam, M.M.; Amir, M.; Akhter, M.; Parvez, S.; Verma, G.; Nainwal, L.M.; Equbal, A.; Anwer, T.; Shaquiquzzaman, M. Heterocyclic Moieties as HDAC Inhibitors: Role in Cancer Therapeutics. Mini Rev. Med. Chem. 2022, 22, 1648–1706. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.D.; Cowley, S.M. The physiological roles of histone deacetylase (HDAC) 1 and 2: Complex co-stars with multiple leading parts. Biochem. Soc. Trans. 2013, 41, 741–749. [Google Scholar] [CrossRef]
- Millard, C.J.; Watson, P.J.; Fairall, L.; Schwabe, J.W.R. Targeting Class I Histone Deacetylases in a “Complex” Environment. Trends Pharmacol. Sci. 2017, 38, 363–377. [Google Scholar] [CrossRef] [PubMed]
- Gadd, M.S.; Testa, A.; Lucas, X.; Chan, K.H.; Chen, W.; Lamont, D.J.; Zengerle, M.; Ciulli, A. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat. Chem. Biol. 2017, 13, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Zengerle, M.; Chan, K.H.; Ciulli, A. Selective Small Molecule Induced Degradation of the BET Bromodomain Protein BRD4. ACS Chem. Biol. 2015, 10, 1770–1777. [Google Scholar] [CrossRef] [PubMed]
- Bondeson, D.P.; Smith, B.E.; Burslem, G.M.; Buhimschi, A.D.; Hines, J.; Jaime-Figueroa, S.; Wang, J.; Hamman, B.D.; Ishchenko, A.; Crews, C.M. Lessons in PROTAC Design from Selective Degradation with a Promiscuous Warhead. Cell Chem. Biol. 2018, 25, 78–87.e75. [Google Scholar] [CrossRef]
- Smith, B.E.; Wang, S.L.; Jaime-Figueroa, S.; Harbin, A.; Wang, J.; Hamman, B.D.; Crews, C.M. Differential PROTAC substrate specificity dictated by orientation of recruited E3 ligase. Nat. Commun. 2019, 10, 131. [Google Scholar] [CrossRef]
- Schiedel, M.; Herp, D.; Hammelmann, S.; Swyter, S.; Lehotzky, A.; Robaa, D.; Oláh, J.; Ovádi, J.; Sippl, W.; Jung, M. Chemically Induced Degradation of Sirtuin 2 (Sirt2) by a Proteolysis Targeting Chimera (PROTAC) Based on Sirtuin Rearranging Ligands (SirReals). J. Med. Chem. 2018, 61, 482–491. [Google Scholar] [CrossRef]
- Yang, K.; Song, Y.; Xie, H.; Wu, H.; Wu, Y.T.; Leisten, E.D.; Tang, W. Development of the first small molecule histone deacetylase 6 (HDAC6) degraders. Bioorg. Med. Chem. Lett. 2018, 28, 2493–2497. [Google Scholar] [CrossRef]
- Xiong, Y.; Donovan, K.A.; Eleuteri, N.A.; Kirmani, N.; Yue, H.; Razov, A.; Krupnick, N.M.; Nowak, R.P.; Fischer, E.S. Chemo-proteomics exploration of HDAC degradability by small molecule degraders. Cell Chem. Biol. 2021, 28, 1514–1527.e1514. [Google Scholar] [CrossRef] [PubMed]
- Smalley, J.P.; Baker, I.M.; Pytel, W.A.; Lin, L.Y.; Bowman, K.J.; Schwabe, J.W.R.; Cowley, S.M.; Hodgkinson, J.T. Optimization of Class I Histone Deacetylase PROTACs Reveals that HDAC1/2 Degradation is Critical to Induce Apoptosis and Cell Arrest in Cancer Cells. J. Med. Chem. 2022, 65, 5642–5659. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Wang, J.; Zhao, L.Y.; Chen, X.; Zheng, G.; Zhang, X.; Liao, D. Discovery of histone deacetylase 3 (HDAC3)-specific PROTACs. Chem. Commun. 2020, 56, 9866–9869. [Google Scholar] [CrossRef] [PubMed]
- Cao, F.; de Weerd, S.; Chen, D.; Zwinderman, M.R.H.; van der Wouden, P.E.; Dekker, F.J. Induced protein degradation of histone deacetylases 3 (HDAC3) by proteolysis targeting chimera (PROTAC). Eur. J. Med. Chem. 2020, 208, 112800. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Geng, A.; Wang, Z.; Wu, C. Efficacy and safety of apatinib combined with liposomal doxorubicin or paclitaxel versus liposomal doxorubicin or paclitaxel monotherapy in patients with recurrent platinum-resistant ovarian cancer. J. Obstet. Gynaecol. Res. 2023, 49, 1611–1619. [Google Scholar] [CrossRef]
- Chotitumnavee, J.; Yamashita, Y.; Takahashi, Y.; Takada, Y.; Iida, T.; Oba, M.; Itoh, Y.; Suzuki, T. Selective degradation of histone deacetylase 8 mediated by a proteolysis targeting chimera (PROTAC). Chem. Commun. 2022, 58, 4635–4638. [Google Scholar] [CrossRef] [PubMed]
- Shen, P.; Wang, Y.; Jia, X.; Xu, P.; Qin, L.; Feng, X.; Li, Z.; Qiu, Z. Dual-target Janus kinase (JAK) inhibitors: Comprehensive review on the JAK-based strategies for treating solid or hematological malignancies and immune-related diseases. Eur. J. Med. Chem. 2022, 239, 114551. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhang, J.; Xu, W.; Wu, Q.; Zeng, R.; Liu, Z.; Tao, W.; Chen, Q.; Wang, Y.; Zhu, W.G. Structure-Based Discovery of Selective Histone Deacetylase 8 Degraders with Potent Anticancer Activity. J. Med. Chem. 2023, 66, 1186–1209. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Chen, D.; Suo, F.; Setroikromo, R.; Quax, W.J.; Dekker, F.J. Discovery of highly potent HDAC8 PROTACs with anti-tumor activity. Bioorg Chem. 2023, 136, 106546. [Google Scholar] [CrossRef]
- Macabuag, N.; Esmieu, W.; Breccia, P.; Jarvis, R.; Blackaby, W.; Lazari, O.; Urbonas, L.; Eznarriaga, M.; Williams, R.; Strijbosch, A.; et al. Developing HDAC4-Selective Protein Degraders to Investigate the Role of HDAC4 in Huntington’s Disease Pathology. J. Med. Chem. 2022, 65, 12445–12459. [Google Scholar] [CrossRef]
- Luckhurst, C.A.; Aziz, O.; Beaumont, V.; Bürli, R.W.; Breccia, P.; Maillard, M.C.; Haughan, A.F.; Lamers, M.; Leonard, P.; Matthews, K.L.; et al. Development and characterization of a CNS-penetrant benzhydryl hydroxamic acid class IIa histone deacetylase inhibitor. Bioorg. Med. Chem. Lett. 2019, 29, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Stott, A.J.; Maillard, M.C.; Beaumont, V.; Allcock, D.; Aziz, O.; Borchers, A.H.; Blackaby, W.; Breccia, P.; Creighton-Gutteridge, G.; Haughan, A.F.; et al. Evaluation of 5-(Trifluoromethyl)-1,2,4-oxadiazole-Based Class IIa HDAC Inhibitors for Huntington’s Disease. ACS Med. Chem. Lett. 2021, 12, 380–388. [Google Scholar] [CrossRef]
- Hook, S.S.; Orian, A.; Cowley, S.M.; Eisenman, R.N. Histone deacetylase 6 binds polyubiquitin through its zinc finger (PAZ domain) and copurifies with deubiquitinating enzymes. Proc. Natl. Acad. Sci. USA 2002, 99, 13425–13430. [Google Scholar] [CrossRef]
- An, Z.; Lv, W.; Su, S.; Wu, W.; Rao, Y. Developing potent PROTACs tools for selective degradation of HDAC6 protein. Protein Cell 2019, 10, 606–609. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Gu, Z.; Lin, S.; Chen, D.; Wang, J.; Zhao, Y.; Li, Y.; Liu, T.; Wang, Y.; Lin, H.; et al. Attenuation of NLRP3 Inflammasome Activation by Indirubin-Derived PROTAC Targeting HDAC6. ACS Chem. Biol. 2021, 16, 2746–2751. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.Y.; Jing, H.; Price, I.R.; Cao, J.; Bai, J.J.; Lin, H. Simultaneous Inhibition of SIRT2 Deacetylase and Defatty-Acylase Activities via a PROTAC Strategy. ACS Med. Chem. Lett. 2020, 11, 2305–2311. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Cueto, M.A.; Asselbergs, F.; Atadja, P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J. Biol. Chem. 2002, 277, 25748–25755. [Google Scholar] [CrossRef] [PubMed]
- Kutil, Z.; Novakova, Z.; Meleshin, M.; Mikesova, J.; Schutkowski, M.; Barinka, C. Histone Deacetylase 11 Is a Fatty-Acid Deacylase. ACS Chem. Biol. 2018, 13, 685–693. [Google Scholar] [CrossRef]
- Bhaskara, S. Histone deacetylase 11 as a key regulator of metabolism and obesity. eBioMedicine 2018, 35, 27–28. [Google Scholar] [CrossRef]
- Son, S.I.; Cao, J.; Zhu, C.L.; Miller, S.P.; Lin, H. Activity-Guided Design of HDAC11-Specific Inhibitors. ACS Chem. Biol. 2019, 14, 1393–1397. [Google Scholar] [CrossRef]
- Banik, D.; Noonepalle, S.; Hadley, M.; Palmer, E.; Gracia-Hernandez, M.; Zevallos-Delgado, C.; Manhas, N.; Simonyan, H.; Young, C.N.; Popratiloff, A.; et al. HDAC6 Plays a Noncanonical Role in the Regulation of Antitumor Immune Responses, Dissemination, and Invasiveness of Breast Cancer. Cancer Res. 2020, 80, 3649–3662. [Google Scholar] [CrossRef] [PubMed]
- Borcoman, E.; Kamal, M.; Marret, G.; Dupain, C.; Castel-Ajgal, Z.; Le Tourneau, C. HDAC Inhibition to Prime Immune Checkpoint Inhibitors. Cancers 2021, 14, 66. [Google Scholar] [CrossRef] [PubMed]
- Glauben, R.; Sonnenberg, E.; Zeitz, M.; Siegmund, B. HDAC inhibitors in models of inflammation-related tumorigenesis. Cancer Lett. 2009, 280, 154–159. [Google Scholar] [CrossRef] [PubMed]
- McIntyre, R.L.; Daniels, E.G.; Molenaars, M.; Houtkooper, R.H.; Janssens, G.E. From molecular promise to preclinical results: HDAC inhibitors in the race for healthy aging drugs. EMBO Mol. Med. 2019, 11, e9854. [Google Scholar] [CrossRef] [PubMed]
- Sanna, M.D.; Guandalini, L.; Romanelli, M.N.; Galeotti, N. The new HDAC1 inhibitor LG325 ameliorates neuropathic pain in a mouse model. Pharmacol. Biochem. Behav. 2017, 160, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Rai, M.; Soragni, E.; Jenssen, K.; Burnett, R.; Herman, D.; Coppola, G.; Geschwind, D.H.; Gottesfeld, J.M.; Pandolfo, M. HDAC inhibitors correct frataxin deficiency in a Friedreich ataxia mouse model. PLoS ONE 2008, 3, e1958. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Pallos, J.; Jacques, V.; Lau, A.; Tang, B.; Cooper, A.; Syed, A.; Purcell, J.; Chen, Y.; Sharma, S.; et al. Histone deacetylase (HDAC) inhibitors targeting HDAC3 and HDAC1 ameliorate polyglutamine-elicited phenotypes in model systems of Huntington’s disease. Neurobiol. Dis. 2012, 46, 351–361. [Google Scholar] [CrossRef]
- Varela, R.B.; Resende, W.R.; Dal-Pont, G.C.; Gava, F.F.; Tye, S.J.; Quevedo, J.; Valvassori, S.S. HDAC inhibitors reverse mania-like behavior and modulate epigenetic regulatory enzymes in an animal model of mania induced by Ouabain. Pharmacol. Biochem. Behav. 2020, 193, 172917. [Google Scholar] [CrossRef]
- Chari, A.; Cho, H.J.; Dhadwal, A.; Morgan, G.; La, L.; Zarychta, K.; Catamero, D.; Florendo, E.; Stevens, N.; Verina, D.; et al. A phase 2 study of panobinostat with lenalidomide and weekly dexamethasone in myeloma. Blood Adv. 2017, 1, 1575–1583. [Google Scholar] [CrossRef]
- Choi, S.W.; Braun, T.; Henig, I.; Gatza, E.; Magenau, J.; Parkin, B.; Pawarode, A.; Riwes, M.; Yanik, G.; Dinarello, C.A.; et al. Vorinostat plus tacrolimus/methotrexate to prevent GVHD after myeloablative conditioning, unrelated donor HCT. Blood 2017, 130, 1760–1767. [Google Scholar] [CrossRef]
- Thomas, A.; Rajan, A.; Szabo, E.; Tomita, Y.; Carter, C.A.; Scepura, B.; Lopez-Chavez, A.; Lee, M.J.; Redon, C.E.; Frosch, A.; et al. A phase I/II trial of belinostat in combination with cisplatin, doxorubicin, and cyclophosphamide in thymic epithelial tumors: A clinical and translational study. Clin. Cancer Res. 2014, 20, 5392–5402. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Sun, Z.; Kuang, P.; Chen, J. Recent progress on HDAC inhibitors with dual targeting capabilities for cancer treatment. Eur. J. Med. Chem. 2020, 208, 112831. [Google Scholar] [CrossRef] [PubMed]
- Almeida, S.; Gao, F.; Coppola, G.; Gao, F.B. Suberoylanilide hydroxamic acid increases progranulin production in iPSC-derived cortical neurons of frontotemporal dementia patients. Neurobiol. Aging 2016, 42, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Witt, O.; Milde, T.; Deubzer, H.E.; Oehme, I.; Witt, R.; Kulozik, A.; Eisenmenger, A.; Abel, U.; Karapanagiotou-Schenkel, I. Phase I/II intra-patient dose escalation study of vorinostat in children with relapsed solid tumor, lymphoma or leukemia. Klin. Padiatr. 2012, 224, 398–403. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, S.; Hase, T.; Shimizu, S.; Ando, M.; Hata, A.; Murakami, H.; Kawakami, T.; Nagase, K.; Yoshimura, K.; Fujiwara, T.; et al. Phase I study of vorinostat with gefitinib in BIM deletion polymorphism/epidermal growth factor receptor mutation double-positive lung cancer. Cancer Sci. 2020, 111, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Abedin, S.M.; Badar, T.; Gauger, K.; Michaelis, L.C.; Runaas, L.; Carlson, K.S.; Murthy, G.G.; Atallah, E. Safety and efficacy of pracinostat in combination with gemtuzumab ozogamicin (PraGO) in patients with relapsed/refractory acute myeloid leukemia. Leuk. Res. 2022, 123, 106984. [Google Scholar] [CrossRef]
- Xue, F.; Cheng, Y.; Xu, L.; Tian, C.; Jiao, H.; Wang, R.; Gao, X. LncRNA NEAT1/miR-129/Bcl-2 signaling axis contributes to HDAC inhibitor tolerance in nasopharyngeal cancer. Aging 2020, 12, 14174–14188. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, Y.; Wang, J.; Zhou, Z.; Cao, S.; Zhang, J. Strategies of Targeting CK2 in Drug Discovery: Challenges, Opportunities, and Emerging Prospects. J. Med. Chem. 2023, 66, 2257–2281. [Google Scholar] [CrossRef] [PubMed]
- Frommel, T.O.; Coon, J.S.; Tsuruo, T.; Roninson, I.B. Variable effects of sodium butyrate on the expression and function of the MDR1 (P-glycoprotein) gene in colon carcinoma cell lines. Int. J. Cancer 1993, 55, 297–302. [Google Scholar] [CrossRef]
- Sun, H.; Li, Y.; Tian, S.; Xu, L.; Hou, T. Assessing the performance of MM/PBSA and MM/GBSA methods. 4. Accuracies of MM/PBSA and MM/GBSA methodologies evaluated by various simulation protocols using PDBbind data set. Phys. Chem. Chem. Phys. 2014, 16, 16719–16729. [Google Scholar] [CrossRef]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Uba, A.I.; Yelekçi, K. Identification of potential isoform-selective histone deacetylase inhibitors for cancer therapy: A combined approach of structure-based virtual screening, ADMET prediction and molecular dynamics simulation assay. J. Biomol. Struct. Dyn. 2018, 36, 3231–3245. [Google Scholar] [CrossRef] [PubMed]
- Venturelli, S.; Berger, A.; Böcker, A.; Busch, C.; Weiland, T.; Noor, S.; Leischner, C.; Schleicher, S.; Mayer, M.; Weiss, T.S.; et al. Resveratrol as a pan-HDAC inhibitor alters the acetylation status of histone [corrected] proteins in human-derived hepatoblastoma cells. PLoS ONE 2013, 8, e73097. [Google Scholar] [CrossRef] [PubMed]
- Urias, B.S.; Pavan, A.R.; Albuquerque, G.R.; Prokopczyk, I.M.; Alves, T.M.F.; de Melo, T.R.F.; Sartori, G.R.; da Silva, J.H.M.; Chin, C.M.; Santos, J.L.D. Optimization of Resveratrol Used as a Scaffold to Design Histone Deacetylase (HDAC-1 and HDAC-2) Inhibitors. Pharmaceuticals 2022, 15, 1260. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Qian, C.; Zhu, Y.; Cai, J.; He, Y.; Li, J.; Wang, T.; Zhu, H.; Li, Z.; Li, W.; et al. Design, synthesis and evaluate of novel dual FGFR1 and HDAC inhibitors bearing an indazole scaffold. Bioorg. Med. Chem. 2018, 26, 747–757. [Google Scholar] [CrossRef]
- Tang, G.; Wong, J.C.; Zhang, W.; Wang, Z.; Zhang, N.; Peng, Z.; Zhang, Z.; Rong, Y.; Li, S.; Zhang, M.; et al. Identification of a novel aminotetralin class of HDAC6 and HDAC8 selective inhibitors. J. Med. Chem. 2014, 57, 8026–8034. [Google Scholar] [CrossRef] [PubMed]
- Bui, H.T.B.; Nguyen, P.H.; Pham, Q.M.; Tran, H.P.; Tran, Q.; Jung, H.; Hong, Q.V.; Nguyen, Q.C.; Nguyen, Q.P.; Le, H.T.; et al. Target Design of Novel Histone Deacetylase 6 Selective Inhibitors with 2-Mercaptoquinazolinone as the Cap Moiety. Molecules 2022, 27, 2204. [Google Scholar] [CrossRef] [PubMed]
- Schäker-Hübner, L.; Warstat, R.; Ahlert, H.; Mishra, P.; Kraft, F.B.; Schliehe-Diecks, J.; Schöler, A.; Borkhardt, A.; Breit, B.; Bhatia, S.; et al. 4-Acyl Pyrrole Capped HDAC Inhibitors: A New Scaffold for Hybrid Inhibitors of BET Proteins and Histone Deacetylases as Antileukemia Drug Leads. J. Med. Chem. 2021, 64, 14620–14646. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.D.; Jiang, Y.J.; Zhou, J.W.; Yu, Q.S.; You, Q.D. Identification of ligand features essential for HDACs inhibitors by pharmacophore modeling. J. Mol. Graph. Model. 2008, 26, 1160–1168. [Google Scholar] [CrossRef]
- Uba, A.I.; Zengin, G. In the quest for histone deacetylase inhibitors: Current trends in the application of multilayered computational methods. Amino Acids 2023, 55, 1709–1726. [Google Scholar] [CrossRef]
- Pai, P.; Kumar, A.; Shetty, M.G.; Kini, S.G.; Krishna, M.B.; Satyamoorthy, K.; Babitha, K.S. Identification of potent HDAC 2 inhibitors using E-pharmacophore modelling, structure-based virtual screening and molecular dynamic simulation. J. Mol. Model. 2022, 28, 119. [Google Scholar] [CrossRef] [PubMed]
- Kumbhar, N.; Nimal, S.; Barale, S.; Kamble, S.; Bavi, R.; Sonawane, K.; Gacche, R. Identification of novel leads as potent inhibitors of HDAC3 using ligand-based pharmacophore modeling and MD simulation. Sci. Rep. 2022, 12, 1712. [Google Scholar] [CrossRef] [PubMed]
- Ganai, S.A.; Abdullah, E.; Rashid, R.; Altaf, M. Combinatorial. Front. Mol. Neurosci. 2017, 10, 357. [Google Scholar] [CrossRef]
- Thangapandian, S.; John, S.; Lee, Y.; Kim, S.; Lee, K.W. Dynamic structure-based pharmacophore model development: A new and effective addition in the histone deacetylase 8 (HDAC8) inhibitor discovery. Int. J. Mol. Sci. 2011, 12, 9440–9462. [Google Scholar] [CrossRef]
- Nencetti, S.; Cuffaro, D.; Nuti, E.; Ciccone, L.; Rossello, A.; Fabbi, M.; Ballante, F.; Ortore, G.; Carbotti, G.; Campelli, F.; et al. Identification of histone deacetylase inhibitors with (arylidene)aminoxy scaffold active in uveal melanoma cell lines. J. Enzyme Inhib. Med. Chem. 2021, 36, 34–47. [Google Scholar] [CrossRef]
- Mehndiratta, S.; Chen, M.C.; Chao, Y.H.; Lee, C.H.; Liou, J.P.; Lai, M.J.; Lee, H.Y. Effect of 3-subsitution of quinolinehydroxamic acids on selectivity of histone deacetylase isoforms. J. Enzyme Inhib. Med. Chem. 2021, 36, 74–84. [Google Scholar] [CrossRef]
- Relitti, N.; Saraswati, A.P.; Chemi, G.; Brindisi, M.; Brogi, S.; Herp, D.; Schmidtkunz, K.; Saccoccia, F.; Ruberti, G.; Ulivieri, C.; et al. Novel quinolone-based potent and selective HDAC6 inhibitors: Synthesis, molecular modeling studies and biological investigation. Eur. J. Med. Chem. 2021, 212, 112998. [Google Scholar] [CrossRef]
- Hu, H.; Chen, F.; Dong, Y.; Li, M.; Xu, S.; Qin, M.; Gong, P. Discovery of Novel c-Mesenchymal-Epithelia transition factor and histone deacetylase dual inhibitors. Eur. J. Med. Chem. 2020, 204, 112651. [Google Scholar] [CrossRef] [PubMed]
- Moffat, D.; Patel, S.; Day, F.; Belfield, A.; Donald, A.; Rowlands, M.; Wibawa, J.; Brotherton, D.; Stimson, L.; Clark, V.; et al. Discovery of 2-(6-{[(6-fluoroquinolin-2-yl)methyl]amino}bicyclo[3.1.0]hex-3-yl)-N-hydroxypyrimidine-5-carboxamide (CHR-3996), a class I selective orally active histone deacetylase inhibitor. J. Med. Chem. 2010, 53, 8663–8678. [Google Scholar] [CrossRef]
- Banerji, U.; van Doorn, L.; Papadatos-Pastos, D.; Kristeleit, R.; Debnam, P.; Tall, M.; Stewart, A.; Raynaud, F.; Garrett, M.D.; Toal, M.; et al. A phase I pharmacokinetic and pharmacodynamic study of CHR-3996, an oral class I selective histone deacetylase inhibitor in refractory solid tumors. Clin. Cancer Res. 2012, 18, 2687–2694. [Google Scholar] [CrossRef]
- Chen, C.; Hou, X.; Wang, G.; Pan, W.; Yang, X.; Zhang, Y.; Fang, H. Design, synthesis and biological evaluation of quinoline derivatives as HDAC class I inhibitors. Eur. J. Med. Chem. 2017, 133, 11–23. [Google Scholar] [CrossRef]
- Zhao, Y.; Yao, Z.; Ren, W.; Yang, X.; Hou, X.; Cao, S.; Fang, H. Design, synthesis and bioactivity evaluations of 8-substituted-quinoline-2-carboxamide derivatives as novel histone deacetylase (HDAC) inhibitors. Bioorg. Med. Chem. 2023, 85, 117242. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Duan, J.; Dang, X.; Yuan, Y.; Wang, Y.; He, X.; Bai, R.; Ye, X.Y.; Xie, T. Design, synthesis and biological evaluation of novel histone deacetylase (HDAC) inhibitors derived from. J. Enzyme Inhib. Med. Chem. 2023, 38, 2195991. [Google Scholar] [CrossRef]
- Zheng, F.; Tang, Q.; Zheng, X.H.; Wu, J.; Huang, H.; Zhang, H.; Hann, S.S. Inactivation of Stat3 and crosstalk of miRNA155-5p and FOXO3a contribute to the induction of IGFBP1 expression by beta-elemene in human lung cancer. Exp. Mol. Med. 2018, 50, 1–14. [Google Scholar] [CrossRef]
- Long, J.; Liu, Z.; Hui, L. Anti-tumor effect and mechanistic study of elemene on pancreatic carcinoma. BMC Complement. Altern. Med. 2019, 19, 133. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Guo, T.; Qu, X.; Hu, X.; Zhang, Y.; Che, X.; Song, H.; Gong, J.; Ma, R.; Li, C.; et al. β-elemene increases the sensitivity of gastric cancer cells to TRAIL by promoting the formation of DISC in lipid rafts. Cell Biol. Int. 2018, 42, 1377–1385. [Google Scholar] [CrossRef]
- Xie, L.; Zhang, J.; Yan, H.; Cai, Y.; Xu, L. β-elemene induced apoptosis and senescence of triple-negative breast cancer cells through IGF1/IGF1R pathway. Tissue Cell 2022, 79, 101914. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Ma, L.; Nong, S.; Wu, Y.; Guo, X.; Pu, J. β-elemene induced anticancer effect in bladder cancer through upregulation of PTEN and suppression of AKT phosphorylation. Oncol. Lett. 2018, 16, 6019–6025. [Google Scholar] [CrossRef] [PubMed]
- Li, C.L.; Chang, L.; Guo, L.; Zhao, D.; Liu, H.B.; Wang, Q.S.; Zhang, P.; Du, W.Z.; Liu, X.; Zhang, H.T.; et al. β-elemene induces caspase-dependent apoptosis in human glioma cells in vitro through the upregulation of Bax and Fas/FasL and downregulation of Bcl-2. Asian Pac. J. Cancer Prev. 2014, 15, 10407–10412. [Google Scholar] [CrossRef]
- Juvale, D.C.; Kulkarni, V.V.; Deokar, H.S.; Wagh, N.K.; Padhye, S.B.; Kulkarni, V.M. 3D-QSAR of histone deacetylase inhibitors: Hydroxamate analogues. Org. Biomol. Chem. 2006, 4, 2858–2868. [Google Scholar] [CrossRef]
- Guo, Y.; Xiao, J.; Guo, Z.; Chu, F.; Cheng, Y.; Wu, S. Exploration of a binding mode of indole amide analogues as potent histone deacetylase inhibitors and 3D-QSAR analyses. Bioorg. Med. Chem. 2005, 13, 5424–5434. [Google Scholar] [CrossRef]
- Banerjee, S.; Adhikari, N.; Amin, S.A.; Jha, T. Structural exploration of tetrahydroisoquinoline derivatives as HDAC8 inhibitors through multi-QSAR modeling study. J. Biomol. Struct. Dyn. 2020, 38, 1551–1564. [Google Scholar] [CrossRef]
- Cao, G.P.; Arooj, M.; Thangapandian, S.; Park, C.; Arulalapperumal, V.; Kim, Y.; Kwon, Y.J.; Kim, H.H.; Suh, J.K.; Lee, K.W. A lazy learning-based QSAR classification study for screening potential histone deacetylase 8 (HDAC8) inhibitors. SAR QSAR Environ. Res. 2015, 26, 397–420. [Google Scholar] [CrossRef] [PubMed]
- Abdul, A.S.; Janish, K.; Samima, K.; Sanjib, D.; Ahmed, Q.I.; Tarun, J.; Shovanlal, G. Binary quantitative activity-activity relationship (QAAR) studies to explore selective HDAC8 inhibitors: In light of mathematical models, DFT-based calculation and molecular dynamic simulation studies. J. Mol. Struct. 2022, 1260, 132833. [Google Scholar]
- Zhao, L.; Fu, L.; Li, G.; Yu, Y.; Wang, J.; Liang, H.; Shu, M.; Lin, Z.; Wang, Y. Three-dimensional quantitative structural-activity relationship and molecular dynamics study of multivariate substituted 4-oxyquinazoline HDAC6 inhibitors. Mol. Divers. 2023, 27, 1123–1140. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.A.; Trivedi, P.; Adhikari, N.; Routholla, G.; Vijayasarathi, D.; Das, S.; Ghosh, B.; Jha, T. Quantitative activity–activity relationship (QAAR) driven design to develop hydroxamate derivatives of pentanoic acids as selective HDAC8 inhibitors: Synthesis, biological evaluation and binding mode of interaction studies. New J. Chem. 2021, 45, 17149–17162. [Google Scholar] [CrossRef]
- Shaker, B.; Ahmad, S.; Lee, J.; Jung, C.; Na, D. In silico methods and tools for drug discovery. Comput. Biol. Med. 2021, 137, 104851. [Google Scholar] [CrossRef] [PubMed]
- Velankar, S.; Burley, S.K.; Kurisu, G.; Hoch, J.C.; Markley, J.L. The Protein Data Bank Archive. Methods Mol. Biol. 2021, 2305, 3–21. [Google Scholar] [CrossRef]
- Lee, H.S.; Park, S.B.; Kim, S.A.; Kwon, S.K.; Cha, H.; Lee, D.Y.; Ro, S.; Cho, J.M.; Song, S.Y. A novel HDAC inhibitor, CG200745, inhibits pancreatic cancer cell growth and overcomes gemcitabine resistance. Sci. Rep. 2017, 7, 41615. [Google Scholar] [CrossRef]
- Ibrahim Uba, A.; Yelekçi, K. Homology modeling of human histone deacetylase 10 and design of potential selective inhibitors. J. Biomol. Struct. Dyn. 2019, 37, 3627–3636. [Google Scholar] [CrossRef]
- Sixto-López, Y.; Gómez-Vidal, J.A.; de Pedro, N.; Bello, M.; Rosales-Hernández, M.C.; Correa-Basurto, J. Hydroxamic acid derivatives as HDAC1, HDAC6 and HDAC8 inhibitors with antiproliferative activity in cancer cell lines. Sci. Rep. 2020, 10, 10462. [Google Scholar] [CrossRef] [PubMed]
- Elmezayen, A.D.; Kemal, Y. Structure-based virtual screening for novel potential selective inhibitors of class IIa histone deacetylases for cancer treatment. Comput. Biol. Chem. 2021, 92, 107491. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yan, W.; Wang, S.; Lu, M.; Yang, H.; Chai, X.; Shi, H.; Zhang, Y.; Jia, Q. Discovery of selective HDAC6 inhibitors based on a multi-layer virtual screening strategy. Comput. Biol. Med. 2023, 160, 107036. [Google Scholar] [CrossRef]
- Peng, Z.; Zhao, Q.; Tian, X.; Lei, T.; Xiang, R.; Chen, L.; Yang, Y. Discovery of Potent and Isoform-selective Histone Deacetylase Inhibitors Using Structure-based Virtual Screening and Biological Evaluation. Mol. Inform. 2022, 41, e2100295. [Google Scholar] [CrossRef]
- Pang, H.; Wu, L.; Tang, Y.; Zhou, G.; Qu, C.; Duan, J.A. Chemical Analysis of the Herbal Medicine Salviae miltiorrhizae Radix et Rhizoma (Danshen). Molecules 2016, 21, 51. [Google Scholar] [CrossRef] [PubMed]
- Shuai, W.; Wang, G.; Zhang, Y.; Bu, F.; Zhang, S.; Miller, D.D.; Li, W.; Ouyang, L.; Wang, Y. Recent Progress on Tubulin Inhibitors with Dual Targeting Capabilities for Cancer Therapy. J. Med. Chem. 2021, 64, 7963–7990. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Qin, J.; Kang, Y.; Zhang, Y.; Ba, M.; Yang, H.; Duan, Y.; Yao, Y. 2-Methoxydiol derivatives as new tubulin and HDAC dual-targeting inhibitors, displaying antitumor and antiangiogenic response. Bioorg. Chem. 2022, 120, 105625. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yan, W.; Yu, Y.; Wang, Y.; Yang, T.; Xue, L.; Yuan, X.; Long, C.; Liu, Z.; Chen, X.; et al. The compound millepachine and its derivatives inhibit tubulin polymerization by irreversibly binding to the colchicine-binding site in β-tubulin. J. Biol. Chem. 2018, 293, 9461–9472. [Google Scholar] [CrossRef]
- Xie, S.; Leng, J.; Zhao, S.; Zhu, L.; Zhang, M.; Ning, M.; Zhao, B.; Kong, L.; Yin, Y. Design and biological evaluation of dual tubulin/HDAC inhibitors based on millepachine for treatment of prostate cancer. Eur. J. Med. Chem. 2024, 268, 116301. [Google Scholar] [CrossRef]
- Ferro, A.; Pantazaka, E.; Athanassopoulos, C.M.; Cuendet, M. Histone deacetylase-based dual targeted inhibition in multiple myeloma. Med. Res. Rev. 2023, 43, 2177–2236. [Google Scholar] [CrossRef]
- Fan, F.; Liu, P.; Bao, R.; Chen, J.; Zhou, M.; Mo, Z.; Ma, Y.; Liu, H.; Zhou, Y.; Cai, X.; et al. A Dual PI3K/HDAC Inhibitor Induces Immunogenic Ferroptosis to Potentiate Cancer Immune Checkpoint Therapy. Cancer Res. 2021, 81, 6233–6245. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Soh, C.K.; Goh, W.H.; Wang, H. Design, Synthesis, and Preclinical Evaluation of Fused Pyrimidine-Based Hydroxamates for the Treatment of Hepatocellular Carcinoma. J. Med. Chem. 2018, 61, 1552–1575. [Google Scholar] [CrossRef] [PubMed]
- Thakur, A.; Tawa, G.J.; Henderson, M.J.; Danchik, C.; Liu, S.; Shah, P.; Wang, A.Q.; Dunn, G.; Kabir, M.; Padilha, E.C.; et al. Design, Synthesis, and Biological Evaluation of Quinazolin-4-one-Based Hydroxamic Acids as Dual PI3K/HDAC Inhibitors. J. Med. Chem. 2020, 63, 4256–4292. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Dai, W.; Chen, X.; Geng, A.; Chen, Y.; Lu, T.; Zhu, Y. Design, synthesis and biological evaluation of 2,3-dihydroimidazo [1,2-c] quinazoline derivatives as novel phosphatidylinositol 3-kinase and histone deacetylase dual inhibitors. RSC Adv. 2017, 7, 52180–52186. [Google Scholar] [CrossRef]
- Tomaselli, D.; Lucidi, A.; Rotili, D.; Mai, A. Epigenetic polypharmacology: A new frontier for epi-drug discovery. Med. Res. Rev. 2020, 40, 190–244. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Lai, F.; Lin, S.; Ji, M.; Zhang, J.; Zhang, Y.; Jin, J.; Fu, R.; Wu, D.; Tian, H.; et al. Design, Synthesis, and Biological Evaluation of 4-Methyl Quinazoline Derivatives as Anticancer Agents Simultaneously Targeting Phosphoinositide 3-Kinases and Histone Deacetylases. J. Med. Chem. 2019, 62, 6992–7014. [Google Scholar] [CrossRef]
- Zhang, K.; Huang, R.; Ji, M.; Lin, S.; Lai, F.; Wu, D.; Tian, H.; Bi, J.; Peng, S.; Hu, J.; et al. Rational design and optimization of novel 4-methyl quinazoline derivatives as PI3K/HDAC dual inhibitors with benzamide as zinc binding moiety for the treatment of acute myeloid leukemia. Eur. J. Med. Chem. 2024, 264, 116015. [Google Scholar] [CrossRef] [PubMed]
- Ballester, P.J. Machine Learning for Molecular Modelling in Drug Design. Biomolecules 2019, 9, 216. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Li, X. A Review on Applications of Computational Methods in Drug Screening and Design. Molecules 2020, 25, 1375. [Google Scholar] [CrossRef]
- Stokes, J.M.; Yang, K.; Swanson, K.; Jin, W.; Cubillos-Ruiz, A.; Donghia, N.M.; MacNair, C.R.; French, S.; Carfrae, L.A.; Bloom-Ackermann, Z.; et al. A Deep Learning Approach to Antibiotic Discovery. Cell 2020, 181, 475–483. [Google Scholar] [CrossRef]
- Rifaioglu, A.S.; Nalbat, E.; Atalay, V.; Martin, M.J.; Cetin-Atalay, R.; Doğan, T. DEEPScreen: High performance drug-target interaction prediction with convolutional neural networks using 2-D structural compound representations. Chem. Sci. 2020, 11, 2531–2557. [Google Scholar] [CrossRef]
- Shimizu, Y.; Yonezawa, T.; Bao, Y.; Sakamoto, J.; Yokogawa, M.; Furuya, T.; Osawa, M.; Ikeda, K. Applying deep learning to iterative screening of medium-sized molecules for protein-protein interaction-targeted drug discovery. Chem. Commun. 2023, 59, 6722–6725. [Google Scholar] [CrossRef] [PubMed]
- Melesina, J.; Simoben, C.V.; Praetorius, L.; Bülbül, E.F.; Robaa, D.; Sippl, W. Strategies To Design Selective Histone Deacetylase Inhibitors. ChemMedChem 2021, 16, 1336–1359. [Google Scholar] [CrossRef] [PubMed]
- Baselious, F.; Hilscher, S.; Robaa, D.; Barinka, C.; Schutkowski, M.; Sippl, W. Comparative Structure-Based Virtual Screening Utilizing Optimized AlphaFold Model Identifies Selective HDAC11 Inhibitor. Int. J. Mol. Sci. 2024, 25, 1358. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A.; Amin, S.A.; Kumar, P.; Jha, T.; Gayen, S. Exploring structural requirements of HDAC10 inhibitors through comparative machine learning approaches. J. Mol. Graph. Model. 2023, 123, 108510. [Google Scholar] [CrossRef] [PubMed]
- Nurani, A.; Yamashita, Y.; Taki, Y.; Takada, Y.; Itoh, Y.; Suzuki, T. Identification of a Histone Deacetylase 8 Inhibitor through Drug Screenings Based on Machine Learning. Chem. Pharm. Bull. 2024, 72, 173–178. [Google Scholar] [CrossRef]
- Li, R.; Tian, Y.; Yang, Z.; Ji, Y.; Ding, J.; Yan, A. Classification models and SAR analysis on HDAC1 inhibitors using machine learning methods. Mol. Divers. 2023, 27, 1037–1051. [Google Scholar] [CrossRef]
- Banerjee, S.; Jana, S.; Jha, T.; Ghosh, B.; Adhikari, N. An assessment of crucial structural contributors of HDAC6 inhibitors through fragment-based non-linear pattern recognition and molecular dynamics simulation approaches. Comput. Biol. Chem. 2024, 110, 108051. [Google Scholar] [CrossRef]
- Sotriffer, C.A.; Sanschagrin, P.; Matter, H.; Klebe, G. SFCscore: Scoring functions for affinity prediction of protein-ligand complexes. Proteins 2008, 73, 395–419. [Google Scholar] [CrossRef]
- Smith, R.D.; Damm-Ganamet, K.L.; Dunbar, J.B.; Ahmed, A.; Chinnaswamy, K.; Delproposto, J.E.; Kubish, G.M.; Tinberg, C.E.; Khare, S.D.; Dou, J.; et al. CSAR Benchmark Exercise 2013: Evaluation of Results from a Combined Computational Protein Design, Docking, and Scoring/Ranking Challenge. J. Chem. Inf. Model. 2016, 56, 1022–1031. [Google Scholar] [CrossRef]
- Spyrakis, F.; Cavasotto, C.N. Open challenges in structure-based virtual screening: Receptor modeling, target flexibility consideration and active site water molecules description. Arch. Biochem. Biophys. 2015, 583, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Poonia, P.; Sharma, M.; Jha, P.; Chopra, M. Pharmacophore-based virtual screening of ZINC database, molecular modeling and designing new derivatives as potential HDAC6 inhibitors. Mol. Divers. 2023, 27, 2053–2071. [Google Scholar] [CrossRef] [PubMed]
- Byvatov, E.; Fechner, U.; Sadowski, J.; Schneider, G. Comparison of support vector machine and artificial neural network systems for drug/nondrug classification. J. Chem. Inf. Comput. Sci. 2003, 43, 1882–1889. [Google Scholar] [CrossRef] [PubMed]
- Zernov, V.V.; Balakin, K.V.; Ivaschenko, A.A.; Savchuk, N.P.; Pletnev, I.V. Drug discovery using support vector machines. The case studies of drug-likeness, agrochemical-likeness, and enzyme inhibition predictions. J. Chem. Inf. Comput. Sci. 2003, 43, 2048–2056. [Google Scholar] [CrossRef] [PubMed]
- Warmuth, M.K.; Liao, J.; Rätsch, G.; Mathieson, M.; Putta, S.; Lemmen, C. Active learning with support vector machines in the drug discovery process. J. Chem. Inf. Comput. Sci. 2003, 43, 667–673. [Google Scholar] [CrossRef]
- Jorissen, R.N.; Gilson, M.K. Virtual screening of molecular databases using a support vector machine. J. Chem. Inf. Model. 2005, 45, 549–561. [Google Scholar] [CrossRef]
- Podolyan, Y.; Walters, M.A.; Karypis, G. Assessing synthetic accessibility of chemical compounds using machine learning methods. J. Chem. Inf. Model. 2010, 50, 979–991. [Google Scholar] [CrossRef] [PubMed]
- Geppert, H.; Horváth, T.; Gärtner, T.; Wrobel, S.; Bajorath, J. Support-vector-machine-based ranking significantly improves the effectiveness of similarity searching using 2D fingerprints and multiple reference compounds. J. Chem. Inf. Model. 2008, 48, 742–746. [Google Scholar] [CrossRef]
- Agarwal, S.; Dugar, D.; Sengupta, S. Ranking chemical structures for drug discovery: A new machine learning approach. J. Chem. Inf. Model. 2010, 50, 716–731. [Google Scholar] [CrossRef]
- Rathke, F.; Hansen, K.; Brefeld, U.; Müller, K.R. StructRank: A new approach for ligand-based virtual screening. J. Chem. Inf. Model. 2011, 51, 83–92. [Google Scholar] [CrossRef]
- Jacob, L.; Hoffmann, B.; Stoven, V.; Vert, J.-P. Virtual screening of GPCRs: An in silico chemogenomics approach. BMC Bioinform. 2008, 9, 363. [Google Scholar] [CrossRef]
- Jacob, L.; Vert, J.P. Protein-ligand interaction prediction: An improved chemogenomics approach. Bioinformatics 2008, 24, 2149–2156. [Google Scholar] [CrossRef]
- Schölkopf, B.; Warmuth, M.K. Learning Theory and Kernel Machines: 16th Annual Conference on Computational Learning Theory and 7th Kernel Workshop, COLT/Kernel 2003, Washington, DC, USA, 24–27 August 2003, Proceedings; Springer: Berlin/Heidelberg, Germany, 2003; Volume 2777. [Google Scholar]
- Vitányi, P. Computational Learning Theory: Second European Conference, EuroCOLT’95, Barcelona, Spain, 13–15 March 1995. Proceedings; Springer Science & Business Media: Berlin/Heidelberg, Germany, 1995; Volume 2. [Google Scholar]
- Breiman, L. Bagging predictors. Mach. Learn. 1996, 24, 123–140. [Google Scholar] [CrossRef]
- Kauffman, G.W.; Jurs, P.C. QSAR and k-nearest neighbor classification analysis of selective cyclooxygenase-2 inhibitors using topologically-based numerical descriptors. J. Chem. Inf. Comput. Sci. 2001, 41, 1553–1560. [Google Scholar] [CrossRef] [PubMed]
- Konovalov, D.A.; Coomans, D.; Deconinck, E.; Heyden, Y.V. Benchmarking of QSAR models for blood-brain barrier permeation. J. Chem. Inf. Model. 2007, 47, 1648–1656. [Google Scholar] [CrossRef]
- Votano, J.R.; Parham, M.; Hall, L.H.; Kier, L.B.; Oloff, S.; Tropsha, A.; Xie, Q.; Tong, W. Three new consensus QSAR models for the prediction of Ames genotoxicity. Mutagenesis 2004, 19, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Zhang, F.; Chu, Q.; Liu, Z. High-performance blob-based iterative three-dimensional reconstruction in electron tomography using multi-GPUs. BMC Bioinform. 2012, 13 (Suppl. S10), S4. [Google Scholar] [CrossRef] [PubMed]
- De Ferrari, L.; Aitken, S.; van Hemert, J.; Goryanin, I. EnzML: Multi-label prediction of enzyme classes using InterPro signatures. BMC Bioinform. 2012, 13, 61. [Google Scholar] [CrossRef]
- Soyguder, S. Intelligent control based on wavelet decomposition and neural network for predicting of human trajectories with a novel vision-based robotic. Expert Syst. Appl. 2011, 38, 13994–14000. [Google Scholar] [CrossRef]
- Haykin, S. Neural Networks: A Comprehensive Foundation; Prentice Hall PTR: Hoboken, NJ, USA, 1998. [Google Scholar]
- Zhang, L.; Zhang, J.; Jiang, Q.; Song, W. Zinc binding groups for histone deacetylase inhibitors. J. Enzyme Inhib. Med. Chem. 2018, 33, 714–721. [Google Scholar] [CrossRef]
- Hu, E.; Dul, E.; Sung, C.M.; Chen, Z.; Kirkpatrick, R.; Zhang, G.F.; Johanson, K.; Liu, R.; Lago, A.; Hofmann, G.; et al. Identification of novel isoform-selective inhibitors within class I histone deacetylases. J. Pharmacol. Exp. Ther. 2003, 307, 720–728. [Google Scholar] [CrossRef] [PubMed]
- Kleinschek, A.; Meyners, C.; Digiorgio, E.; Brancolini, C.; Meyer-Almes, F.J. Potent and Selective Non-hydroxamate Histone Deacetylase 8 Inhibitors. ChemMedChem 2016, 11, 2598–2606. [Google Scholar] [CrossRef]
- Ononye, S.N.; VanHeyst, M.D.; Oblak, E.Z.; Zhou, W.; Ammar, M.; Anderson, A.C.; Wright, D.L. Tropolones as lead-like natural products: The development of potent and selective histone deacetylase inhibitors. ACS Med. Chem. Lett. 2013, 4, 757–761. [Google Scholar] [CrossRef] [PubMed]
- Patil, V.; Sodji, Q.H.; Kornacki, J.R.; Mrksich, M.; Oyelere, A.K. 3-Hydroxypyridin-2-thione as novel zinc binding group for selective histone deacetylase inhibition. J. Med. Chem. 2013, 56, 3492–3506. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Stowe, R.L.; Pinello, C.E.; Tian, G.; Madoux, F.; Li, D.; Zhao, L.Y.; Li, J.L.; Ma, H.; Hodder, P.; et al. Identification of histone deacetylase inhibitors with benzoylhydrazide scaffold that selectively inhibit class I histone deacetylases. Chem. Biol. 2015, 22, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Lobera, M.; Madauss, K.P.; Pohlhaus, D.T.; Wright, Q.G.; Trocha, M.; Schmidt, D.R.; Baloglu, E.; Trump, R.P.; Head, M.S.; Hofmann, G.A.; et al. Selective class IIa histone deacetylase inhibition via a nonchelating zinc-binding group. Nat. Chem. Biol. 2013, 9, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Maolanon, A.R.; Madsen, A.S.; Olsen, C.A. Innovative Strategies for Selective Inhibition of Histone Deacetylases. Cell Chem. Biol. 2016, 23, 759–768. [Google Scholar] [CrossRef] [PubMed]
- Abdizadeh, T.; Ghodsi, R.; Hadizadeh, F. 3D-QSAR (CoMFA, CoMSIA) and Molecular Docking Studies on Histone Deacetylase 1 Selective Inhibitors. Recent. Pat. Anticancer. Drug Discov. 2017, 12, 365–383. [Google Scholar] [CrossRef]
- Cao, F.; Zwinderman, M.R.H.; Dekker, F.J. The Process and Strategy for Developing Selective Histone Deacetylase 3 Inhibitors. Molecules 2018, 23, 551. [Google Scholar] [CrossRef]
- Suzuki, T.; Kasuya, Y.; Itoh, Y.; Ota, Y.; Zhan, P.; Asamitsu, K.; Nakagawa, H.; Okamoto, T.; Miyata, N. Identification of highly selective and potent histone deacetylase 3 inhibitors using click chemistry-based combinatorial fragment assembly. PLoS ONE 2013, 8, e68669. [Google Scholar] [CrossRef]
- Ruzic, D.; Djokovic, N.; Nikolic, K. Fragment-Based Drug Design of Selective HDAC6 Inhibitors. Methods Mol. Biol. 2021, 2266, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Moi, D.; Citarella, A.; Bonanni, D.; Pinzi, L.; Passarella, D.; Silvani, A.; Giannini, C.; Rastelli, G. Synthesis of potent and selective HDAC6 inhibitors led to unexpected opening of a quinazoline ring. RSC Adv. 2022, 12, 11548–11556. [Google Scholar] [CrossRef] [PubMed]
- Maruca, A.; Moraca, F.; Rocca, R.; Molisani, F.; Alcaro, F.; Gidaro, M.C.; Alcaro, S.; Costa, G.; Ortuso, F. Chemoinformatic Database Building and in Silico Hit-Identification of Potential Multi-Targeting Bioactive Compounds Extracted from Mushroom Species. Molecules 2017, 22, 1571. [Google Scholar] [CrossRef] [PubMed]
- Barbaraci, C.; di Giacomo, V.; Maruca, A.; Patamia, V.; Rocca, R.; Dichiara, M.; Di Rienzo, A.; Cacciatore, I.; Cataldi, A.; Balaha, M.; et al. Discovery of first novel sigma/HDACi dual-ligands with a potent in vitro antiproliferative activity. Bioorg. Chem. 2023, 140, 106794. [Google Scholar] [CrossRef]
- Patel, V.K.; Shirbhate, E.; Tiwari, P.; Kore, R.; Veerasamy, R.; Mishra, A.; Rajak, H. Multi-targeted HDAC Inhibitors as Anticancer Agents: Current Status and Future Prospective. Curr. Med. Chem. 2023, 30, 2762–2795. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.X.; Huang, J.; Tian, X.Y.; Liu, Y.H.; Jia, M.Q.; Wang, W.; Jin, C.Y.; Song, J.; Zhang, S.Y. A review of progress in o-aminobenzamide-based HDAC inhibitors with dual targeting capabilities for cancer therapy. Eur. J. Med. Chem. 2023, 259, 115673. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.W.; Shou, D.; Huang, R.; Khuc, T.; Dai, S.; Zheng, W.; Klumpp-Thomas, C.; Xia, M. Identification of HDAC Inhibitors Using a Cell-Based HDAC I/II Assay. J. Biomol. Screen. 2016, 21, 643–652. [Google Scholar] [CrossRef]
- Mehdi, O.; Françoise, S.; Sofia, C.L.; Urs, G.; Kevin, Z.; Bernard, S.; Igor, S.; Anabela, C.D.; Dominique, L.; Eric, M.; et al. HDAC gene expression in pancreatic tumor cell lines following treatment with the HDAC inhibitors panobinostat (LBH589) and trichostatine (TSA). Pancreatology 2012, 12, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Weiss, U.; Möller, M.; Husseini, S.A.; Manderscheid, C.; Häusler, J.; Geisslinger, G.; Niederberger, E. Inhibition of HDAC Enzymes Contributes to Differential Expression of Pro-Inflammatory Proteins in the TLR-4 Signaling Cascade. Int. J. Mol. Sci. 2020, 21, 8943. [Google Scholar] [CrossRef]
- Lopez, G.; Song, Y.; Lam, R.; Ruder, D.; Creighton, C.J.; Bid, H.K.; Bill, K.L.; Bolshakov, S.; Zhang, X.; Lev, D.; et al. HDAC Inhibition for the Treatment of Epithelioid Sarcoma: Novel Cross Talk Between Epigenetic Components. Mol. Cancer Res. 2016, 14, 35–43. [Google Scholar] [CrossRef]
- Yu, W.C.; Yeh, T.Y.; Ye, C.H.; Chong, P.C.T.; Ho, Y.H.; So, D.K.; Yap, K.Y.; Peng, G.R.; Shao, C.H.; Jagtap, A.D.; et al. Discovery of HDAC6, HDAC8, and 6/8 Inhibitors and Development of Cell-Based Drug Screening Models for the Treatment of TGF-β-Induced Idiopathic Pulmonary Fibrosis. J. Med. Chem. 2023, 66, 10528–10557. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Jeffers, M.; Kumar, S.; Hackett, C.; Boldog, F.; Khramtsov, N.; Qian, X.; Mills, E.; Berghs, S.C.; Carey, N.; et al. Determination of the class and isoform selectivity of small-molecule histone deacetylase inhibitors. Biochem. J. 2008, 409, 581–589. [Google Scholar] [CrossRef]
- Butler, K.V.; Kalin, J.; Brochier, C.; Vistoli, G.; Langley, B.; Kozikowski, A.P. Rational design and simple chemistry yield a superior, neuroprotective HDAC6 inhibitor, tubastatin A. J. Am. Chem. Soc. 2010, 132, 10842–10846. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Altamura, S.; De Francesco, R.; Gallinari, P.; Lahm, A.; Neddermann, P.; Rowley, M.; Serafini, S.; Steinkühler, C. Probing the elusive catalytic activity of vertebrate class IIa histone deacetylases. Bioorg. Med. Chem. Lett. 2008, 18, 1814–1819. [Google Scholar] [CrossRef] [PubMed]
- Estiu, G.; West, N.; Mazitschek, R.; Greenberg, E.; Bradner, J.E.; Wiest, O. On the inhibition of histone deacetylase 8. Bioorg. Med. Chem. 2010, 18, 4103–4110. [Google Scholar] [CrossRef]
- Padige, G.; Negmeldin, A.T.; Pflum, M.K. Development of an ELISA-Based HDAC Activity Assay for Characterization of Isoform-Selective Inhibitors. J. Biomol. Screen. 2015, 20, 1277–1285. [Google Scholar] [CrossRef]


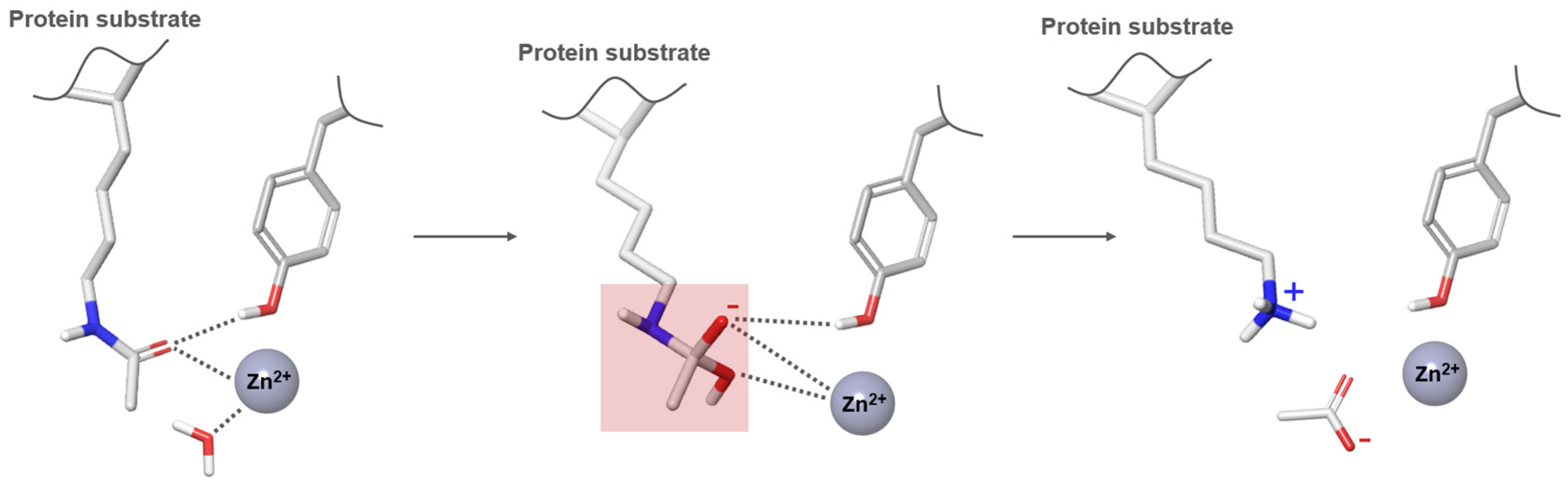




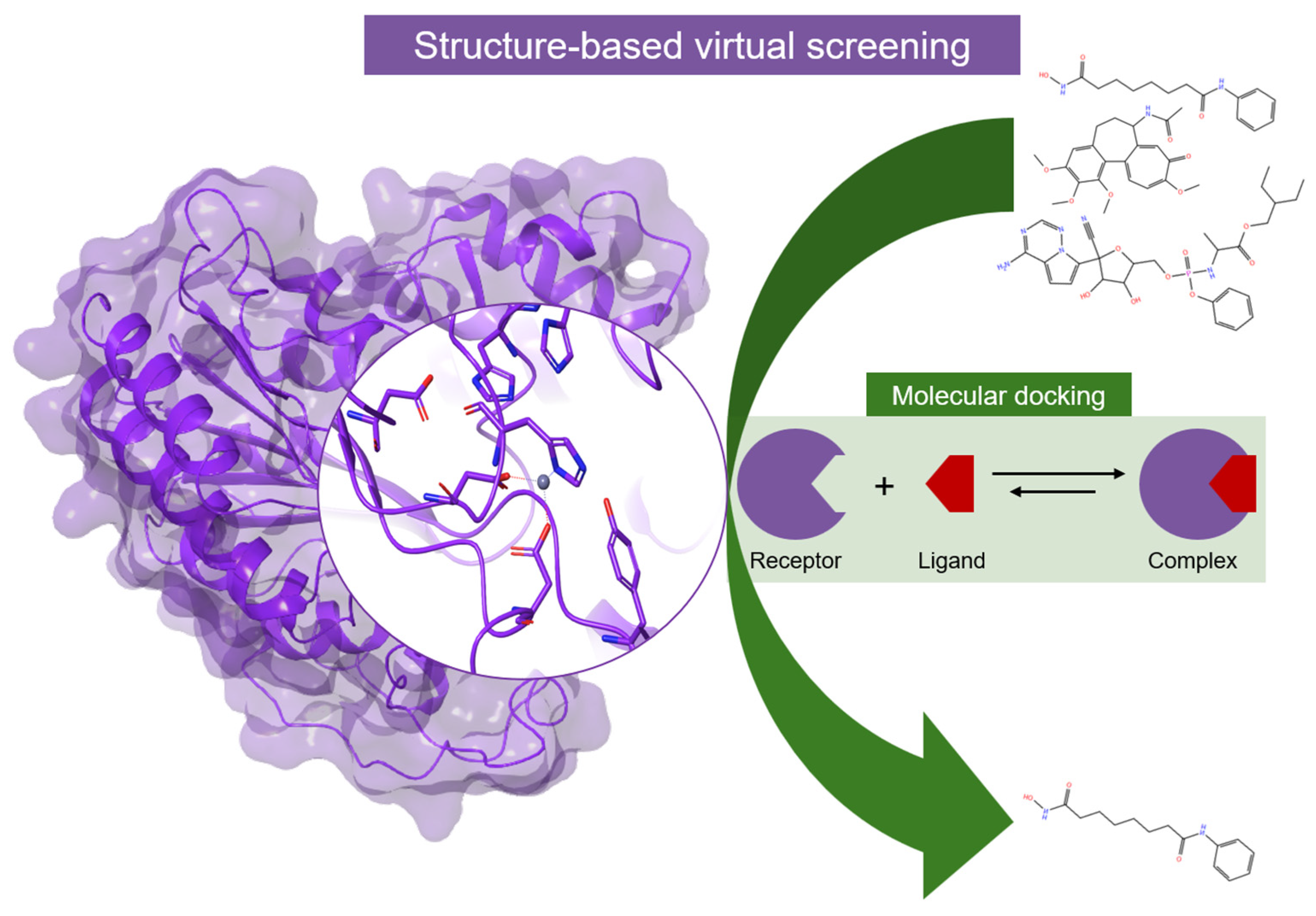

| Class I | Zn2+-Dependent | |
|---|---|---|
| Isoform | Disease | |
| HDAC1, HDAC2, HDAC3, HDAC8 | Cancer (prostate, gastric, colorectal, Hodgkin lymphoma, lung, liver, acute lymphoblastic leukemia, breast, neuroblastoma); Neurological diseases (Huntington’s disease, Amyotrophic Lateral Sclerosis, Alzheimer’s Disease); Metabolic diseases (Diabetes, obesity); Cardiovascular diseases | |
| Class II | Zn2+-Dependent | |
| Isoform | Disease | |
| HDAC4, HDAC5, HDAC6, HDAC7, HDAC9, HDAC10 | Cancer (esophageal, colon, Hodgkin lymphoma, lung, acute lymphoblastic leukemia, breast, medulloblastoma); Neurological diseases (Huntington’s disease, Parkinson’s disease, Amyotrophic Lateral Sclerosis, Alzheimer’s Disease); Cardiovascular diseases | |
| Class IV | Zn2+-Dependent | |
| Isoform | Disease | |
| HDAC11 | Cancer (breast, renal, liver); Neurological diseases (Multiple Sclerosis); Cardiovascular diseases | |
| Class III | NAD+-dependent | |
| Isoform | Disease | |
| SIRT1, SIRT2, SIRT3, SIRT4, SIRT5, SIRT6, SIRT7 | Cancer (breast, pancreatic, colon, glioma); Neurological disease (Multiple Sclerosis); Metabolic diseases (Diabetes, obesity); Cardiovascular diseases | |
| Class I | Zn2+-Dependent | |
|---|---|---|
| Isoform | Role in Cancer Biology | Expression in Cancer |
| HDAC1 | (−) Apoptosis, (+) proliferation | Overexpressed in prostate (hormone-refractory), gastric A, colorectal, breast B, Hodgkin lymphoma, lung A, and liver A cancer |
| HDAC2 | (−) Apoptosis, (+) proliferation, (+) aneuploidy | Overexpressed in colorectal A, gastric A, prostvate A, Hodgkin lymphoma, acute lymphoblastic leukemia |
| HDAC3 | (−) Differentation, (+) proliferation | Overexpressed in lung, gastric A, breast AB, Hodgkin lymphoma, acute lymphoblastic leukemia Downregulated in liver cancer |
| HDAC8 | (−) Differentation, (+) proliferation | Overexpressed in neuroblastoma |
| Class II | Zn2+-Dependent | |
| Isoform | Role in Cancer Biology | Expression in Cancer |
| HDAC4 | (−) Differentation, (+) Angiogenesis | Overexpressed in esophageal cancer |
| HDAC5 | (−) Differentation, (−) Migration | Overexpressed in medulloblastoma Downregulated in lung, colon cancer and acute myeloid leukaemia |
| HDAC6 | (+) Migration | Overexpressed in breast cancer Downregulated in lung cancer |
| HDAC7 | (+) Angiogenesis | Overexpressed in acute lymphoblastic leukemia Downregulated in lung cancer |
| HDAC9 | (+) Angiogenesis | Overexpressed in medulloblastoma |
| HDAC10 | (+) Angiogenesis | Overexpressed in lung cancer |
| Class IV | Zn2+-Dependent | |
| Isoform | Role in Cancer Biology | Expression in Cancer |
| HDAC11 | Overexpressed in breast, renal and liver cancer | |
| Class I | Zn2+-Dependent | ||||
|---|---|---|---|---|---|
| Isoform | Size (aa) | Zn Coordination | Cellular Distribution | Complex | Chromosome Location |
| HDAC1 | 482 | His140 His141 Asp176 Asp264 | Nuclear | Sin3, NURD | 1p34 |
| HDAC2 | 488 | His141 His142 Asp177 Asp265 | Nuclear | Sin3, NURD | 6q21 |
| HDAC3 | 428 | His134 His135 Asp170 Asp259 | Nuclear | NCOR1/NCOR2-GPS2-TBL1X | 5q31 |
| HDAC8 | 377 | His142 His143 Asp178 Asp267 | Nuclear | Xq13 | |
| Class II | Zn2+-Dependent | ||||
| Isoform | Size (aa) | Zn Coordination | Cellular Distribution | Complex | Chromosome Location |
| HDAC4 | 1084 | His802 His803 Asp840 Asp934 | Nuclear, cytoplasm | NCOR1/NCOR2 | q37.2 |
| HDAC5 | 1122 | Nuclear, cytoplasm | 17q21 | ||
| HDAC6 | 1215 | H610 H611 Asp649 Asp742 | Nuclear, cytoplasm | Xp11.22–23 | |
| HDAC7 | 952 | His669 His670 Asp707 Asp801 | Nuclear, cytoplasm | NCOR1/NCOR2 | 12q13.1 |
| HDAC9 | 1011 | Nuclear, cytoplasm | p21–p15 | ||
| HDAC10 | 669 | Nuclear, cytoplasm | NCOR2 | 22q13.31 | |
| Class IV | Zn2+-Dependent | ||||
| Isoform | Size (aa) | Zn Coordination | Cellular Distribution | Complex | Chromosome Location |
| HDAC11 | 347 | Nuclear | - | - | |
| Class III (SIRT) | NAD+-Dependent | ||||
| Isoform | Size (aa) | Cellular Distribution | Complex | Chromosome Location | |
| SIRT1 | 747 | Nuclear, cytoplasm | eNoSC | ||
| SIRT2 | 389 | Nuclear, cytoplasm | |||
| SIRT3 | 399 | Mitochondria | FoxO3A | ||
| SIRT4 | 314 | Mitochondria | |||
| SIRT5 | 310 | Nuclear, cytoplasm, mitochondria | 6p23 | ||
| SIRT6 | 355 | Nucleas, endoplasmic reticulum | |||
| SIRT7 | 400 | Nuclear, cytoplasm | 17q25.3 | ||
| Omics | Analysis | Detecting |
|---|---|---|
| Chemoproteome | MS, beads MS, MudPIT | Protein/HDACi interaction |
| Epigenome | ChIP-seq, ChIP-qPCR, ChIP-chip, DNase-seq, MNase-seq, ATAC-seq, MBD-seq, RNA-seq, NA-seq, HT-FAIRE | Histone modification and chromatin accessibility |
| Acetylome | Protein modification | |
| Transcriptome | Microarray, miRNA microarray, miRNA-seq, mRNA-seq, splicing-sensitive microarray, TempO-seq, GRO-seq, ChIP-seq | Gene expression |
| Proteome | LC-MS/MS, SILAC, HSMS, MS acetylome | Protein expression |
| Metabolome | MS metabolomics, NMR, LC/GC-MS/MS | Metabolic physiology |
| Class | Inhibitor | 2D Structures | Selectivity | Disease |
|---|---|---|---|---|
| Hydroxamic acids | Vorinostat |  | Pan | Cutaneous T-cell lymphoma (Approved) Alzheimer’s disease (Phase I) |
| Trichostatin A |  | Pan | Preclinical use | |
| Belinostat |  | Pan | Peripheral T-cell lymphoma (Approved) | |
| Panobinostat |  | Pan | Multiple myeloma (Approved) | |
| Givinostat |  | Pan | Relapsed leukemia Multiple myeloma (Phase II) | |
| Resminostat |  | Pan | Hepatocellular carcinoma (Phase I and II) | |
| Abexinostat |  | Pan | B-cell lymphoma (Phase II) | |
| Quisinostat |  | Pan | Multiple myeloma (Phase I) | |
| Rocilinostat |  | II | Multiple myeloma (Phase I) | |
| Pracinostat |  | I, II, IV | Prostate cancer (Phase II) | |
| CHR-3996 |  | I | Advanced/metastatic solid tumors refractory to standard therapy (Phase I) | |
| Benzamides | Entinostat |  | I | Breast cancer, Hodgkin’s lymphoma, non-small cell lung cancer (Phase II and Phase III) |
| Tacedinaline |  | I | Hormone receptor-positive breast cancer, Non-small cell lung cancer and pancreatic cancer (Phase III) | |
| 4SC202 |  | I | Advanced hematological malignancies (Phase I) | |
| Mocetinostat | 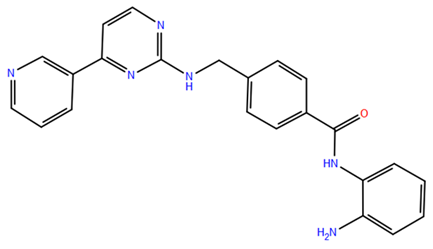 | I, IV | Hodgkin’s lymphoma (Phase II) | |
| Short-chain fatty acids | Valproic acid |  | I, IIa | Epilepsia, bipolar disorders, and migraine (Approved) |
| Butyric acid |  | I, II | Multiple conditions (Phase II) | |
| Phenylbutyric acid |  | I, II | Multiple conditions (Phase I) | |
| Cyclic tetrapeptides | Romidepsin |  | I | Cutaneous T-cell lymphoma (Approved) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Curcio, A.; Rocca, R.; Alcaro, S.; Artese, A. The Histone Deacetylase Family: Structural Features and Application of Combined Computational Methods. Pharmaceuticals 2024, 17, 620. https://doi.org/10.3390/ph17050620
Curcio A, Rocca R, Alcaro S, Artese A. The Histone Deacetylase Family: Structural Features and Application of Combined Computational Methods. Pharmaceuticals. 2024; 17(5):620. https://doi.org/10.3390/ph17050620
Chicago/Turabian StyleCurcio, Antonio, Roberta Rocca, Stefano Alcaro, and Anna Artese. 2024. "The Histone Deacetylase Family: Structural Features and Application of Combined Computational Methods" Pharmaceuticals 17, no. 5: 620. https://doi.org/10.3390/ph17050620






