Evaluation of Serial Procalcitonin Levels for the Optimization of Antibiotic Use in Non-Critically Ill COVID-19 Patients
Abstract
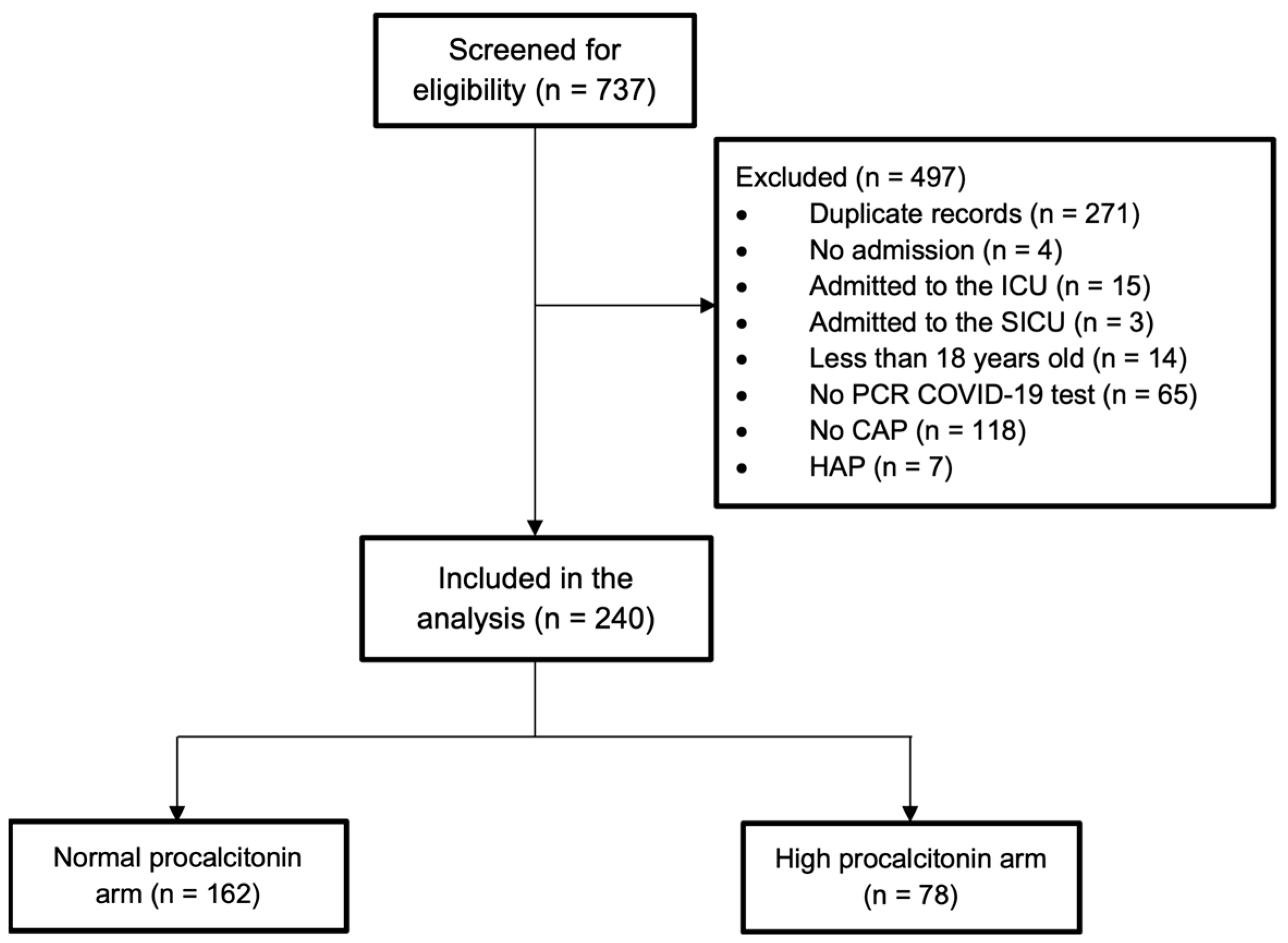
:1. Introduction
2. Results
2.1. Baseline Characteristics
2.2. Study Outcomes
3. Discussion
4. Materials and Methods
4.1. Study Design and Setting
4.2. Study Procedures and Participants
4.3. Study Outcomes
4.4. Ethical Consideration
4.5. Sample Size Calculation
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- The Burden of Antimicrobial Resistance (AMR) in Saudi Arabia. Available online: https://www.healthdata.org/sites/default/files/files/Projects/GRAM/Saudi_Arabia_0.pdf (accessed on 17 April 2024).
- Dupuy, A.M.; Philippart, F.; Péan, Y.; Lasocki, S.; Charles, P.-E.; Chalumeau, M.; Claessens, Y.-E.; Quenot, J.-P.; Guen, C.G.-L.; Ruiz, S.; et al. Role of biomarkers in the management of antibiotic therapy: An expert panel review: I—currently available biomarkers for clinical use in acute infections. Ann. Intensive Care 2013, 3, 22. [Google Scholar] [CrossRef] [PubMed]
- Azzini, A.M.; Dorizzi, R.M.; Sette, P.; Vecchi, M.; Coledan, I.; Righi, E.; Tacconelli, E. A 2020 review on the role of procalcitonin in different clinical settings: An update conducted with the tools of the Evidence-Based Laboratory Medicine. Ann. Transl. Med. 2020, 8, 610. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, S.; Hochreiter, M.; Koehler, T.; Schweiger, A.M.; Bein, B.; Keck, F.S.; von Spiegel, T. Procalcitonin (PCT)-guided algorithm reduces length of antibiotic treatment in surgical intensive care patients with severe sepsis: Results of a prospective randomized study. Langenbecks Arch. Surg. 2009, 394, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Kopterides, P.; Siempos, I.I.; Tsangaris, I.; Tsantes, A.; Armaganidis, A. Procalcitonin-guided algorithms of antibiotic therapy in the intensive care unit: A systematic review and meta-analysis of randomized controlled trials. Crit. Care Med. 2010, 38, 2229–2241. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Sharma, L.; Chang, D. Pathophysiology and clinical management of coronavirus disease (COVID-19): A mini-review. Front. Immunol. 2023, 14, 1116131. [Google Scholar] [CrossRef] [PubMed]
- Marik, P.E.; Iglesias, J.; Varon, J.; Kory, P. A scoping review of the pathophysiology of COVID-19. Int. J. Immunopathol. Pharmacol. 2021, 35, 205873842110480. [Google Scholar] [CrossRef] [PubMed]
- Tziolos, N.R.; Ioannou, P.; Baliou, S.; Kofteridis, D.P. Long COVID-19 pathophysiology: What do we know so far? Microorganisms 2023, 11, 2458. [Google Scholar] [CrossRef] [PubMed]
- Abboud, H.; Abboud, F.Z.; Kharbouch, H.; Arkha, Y.; El Abbadi, N.; El Ouahabi, A. COVID-19 and SARS-CoV-2 infection: Pathophysiology and clinical effects on the nervous system. World Neurosurg. 2020, 140, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Arabi, Y.M.; Al-Omari, A.; Mandourah, Y.; Al-Hameed, F.; Sindi, A.A.; Alraddadi, B.; Shalhoub, S.; Almotairi, A.; Al Khatib, K.; Abdulmomen, A.; et al. Critically Ill Patients with the Middle East Respiratory Syndrome: A Multicenter Retrospective Cohort Study. Crit. Care Med. 2017, 45, 1683–1695. [Google Scholar] [CrossRef] [PubMed]
- Rice, T.W.; Rubinson, L.; Uyeki, T.M.; Vaughn, F.L.; John, B.B.P.; Miller, R.R.I.; Higgs, E.M.; Randolph, A.G.M.; Smoot, B.E.; Thompson, B.T.; et al. Critical Illness from 2009 Pandemic Influenza A (H1N1) Virus and Bacterial Co-Infection in the United States. Crit. Care Med. 2012, 40, 1487. [Google Scholar] [CrossRef] [PubMed]
- Liu, V.X.; Fielding-Singh, V.; Greene, J.D.; Baker, J.M.; Iwashyna, T.J.; Bhattacharya, J.; Escobar, G.J. The Timing of Early Antibiotics and Hospital Mortality in Sepsis. Am. J. Respir. Crit. Care Med. 2017, 196, 856–863. [Google Scholar] [CrossRef] [PubMed]
- Alshaikh, F.S.; Godman, B.; Sindi, O.N.; Seaton, R.A.; Kurdi, A. Prevalence of bacterial coinfection and patterns of antibiotics prescribing in patients with COVID-19: A systematic review and meta-analysis. PLoS ONE 2022, 17, e0272375. [Google Scholar] [CrossRef] [PubMed]
- Shiferaw, B.; Bekele, E.; Kumar, K.; Boutin, A.; Frieri, M. The role of procalcitonin as a biomarker in sepsis. J. Infect. Dis. Epidemiol. 2016, 2, 10-23937. [Google Scholar] [CrossRef]
- Pal, S.; Sengupta, S.; Lahiri, S.; Ghosh, A.; Bhowmick, K. Role of biomarkers in prognostication of moderate and severe COVID-19 cases. J. Fam. Med. Prim. Care 2023, 12, 3186–3193. [Google Scholar] [CrossRef] [PubMed]
- Delévaux, I.; André, M.; Colombier, M.; Albuisson, E.; Meylheuc, F.; Bégue, R.-J.; Piette, J.-C.; Aumaître, O. Can procalcitonin measurement help in differentiating between bacterial infection and other kinds of inflammatory processes? Ann. Rheum. Dis. 2003, 62, 337–340. [Google Scholar] [CrossRef] [PubMed]
- Nobre, V.; Harbarth, S.; Graf, J.D.; Rohner, P.; Pugin, J. Use of procalcitonin to shorten antibiotic treatment duration in septic patients: A randomized trial. Am. J. Respir. Crit. Care Med. 2008, 177, 498–505. [Google Scholar] [CrossRef] [PubMed]
- Bouadma, L.; Luyt, C.E.; Tubach, F.; Cracco, C.; Alvarez, A.; Schwebel, C.; Schortgen, F.; Lasocki, S.; Veber, B.; Dehoux, M.; et al. Use of procalcitonin to reduce patients’ exposure to antibiotics in intensive care units (PRORATA trial): A multicentre randomised controlled trial. Lancet 2010, 375, 463–474. [Google Scholar] [CrossRef] [PubMed]
- Schuetz, P.; Christ-Crain, M.; Thomann, R.; Falconnier, C.; Wolbers, M.; Widmer, I.; Neidert, S.; Fricker, T.; Blum, C.; Schild, U.; et al. Effect of procalcitonin-based guidelines vs standard guidelines on antibiotic use in lower respiratory tract infections: The ProHOSP randomized controlled trial. JAMA 2009, 302, 1059–1066. [Google Scholar] [CrossRef] [PubMed]
- Briel, M.; Schuetz, P.; Mueller, B.; Young, J.; Schild, U.; Nusbaumer, C.; Périat, P.; Bucher, H.C.; Christ-Crain, M. Procalcitonin-guided antibiotic use vs. a standard approach for acute respiratory tract infections in primary care. Arch. Intern. Med. 2008, 168, 2000–2007. [Google Scholar] [CrossRef] [PubMed]
- Christ-Crain, M.; Stolz, D.; Bingisser, R.; Müller, C.; Miedinger, D.; Huber, P.R.; Zimmerli, W.; Harbarth, S.; Tamm, M.; Müller, B. Procalcitonin guidance of antibiotic therapy in community-acquired pneumonia: A randomized trial. Am. J. Respir. Crit. Care Med. 2006, 174, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Christ-Crain, M.; Jaccard-Stolz, D.; Bingisser, R.; Gencay, M.M.; Huber, P.R.; Tamm, M.; Müller, B. Effect of procalcitonin-guided treatment on antibiotic use and outcome in lower respiratory tract infections: Cluster-randomised, single-blinded intervention trial. Lancet 2004, 363, 600–607. [Google Scholar] [CrossRef] [PubMed]
- Müller, B.; Becker, K.L.; Schächinger, H.; Rickenbacher, P.R.; Huber, P.R.; Zimmerli, W.; Ritz, R. Calcitonin precursors are reliable markers of sepsis in a medical intensive care unit. Crit. Care Med. 2000, 28, 977–983. [Google Scholar] [CrossRef] [PubMed]
- So, W.; Simon, M.S.; Choi, J.J.; Wang, T.Z.; Williams, S.C.; Chua, J.; Kubin, C.J. Characteristics of procalcitonin in hospitalized COVID-19 patients and clinical outcomes of antibiotic use stratified by procalcitonin levels. Intern. Emerg. Med. 2022, 17, 1405–1412. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Powers, H.R.; Craver, E.C.; Nazareno, M.D.; Yarrarapu, S.N.S.; Sanghavi, D.K. Antibiotic stewardship: Early discontinuation of antibiotics based on procalcitonin level in COVID-19 pneumonia. J. Clin. Pharm. Ther. 2022, 47, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, R.; Urgelés, S.; Salgado, M.; Rodríguez, A.; Reyes, L.F.; Fuentes, Y.V.; Serrano, C.C.; Caceres, E.L.; Bodí, M.; Martín-Loeches, I.; et al. Negative predictive value of procalcitonin to rule out bacterial respiratory co-infection in critical COVID-19 patients. J. Infect. 2022, 85, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Han, C.; Pei, S.; Yin, M.; Chen, X. Procalcitonin levels in COVID-19 patients. Int. J. Antimicrob. Agents 2020, 56, 106051. [Google Scholar] [CrossRef] [PubMed]
- Fridkin, S.; Baggs, J.; Fagan, R.; Magill, S.; Pollack, L.A.; Malpiedi, P.; Slayton, R.; Khader, K.; Rubin, M.A.; Jones, M.; et al. Vital signs: Improving antibiotic use among hospitalized patients. MMWR Morb. Mortal Wkly. Rep. 2014, 63, 194–200. [Google Scholar] [PubMed]
- Vâţă, A.; Roşu, F.M.; Dorneanu, O.S.; Lehaci, A.E.; Luca, C.M.; Loghin, I.I.; Miftode, I.D.; Luca, C.M.; Miftode, E.G. Antibiotic Usage in the COVID-19 Intensive Care Unit of an Infectious Diseases Hospital from Nord-Eastern Romania. Medicina 2023, 59, 645. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Kang, M.; Shin, D.H.; Jung, J.; Choi, S.J.; Kim, N.-H.; Moon, S.M.; Song, K.-H.; Kim, E.S.; Jung, J.; et al. Antibiotic Prescription in Patients with Coronavirus Disease 2019: Analysis of National Health Insurance System Data in the Republic of Korea. J. Korean Med. Sci. 2023, 38, e189. [Google Scholar] [CrossRef] [PubMed]
- Yunus, I.; Fasih, A.; Wang, Y. The use of procalcitonin in the determination of severity of sepsis, patient outcomes and infection characteristics. PLoS ONE 2018, 13, e0206527. [Google Scholar] [CrossRef] [PubMed]
- Vijayan, A.L.; Vanimaya Ravindran, S.; Saikant, R.; Lakshmi, S.; Kartik, R.; Manoj, G. Procalcitonin: A promising diagnostic marker for sepsis and antibiotic therapy. J. Intensive Care 2017, 5, 51. [Google Scholar] [CrossRef] [PubMed]
- Schuetz, P.; Birkhahn, R.; Sherwin, R.; Jones, A.E.; Singer, A.; Kline, J.A.; Runyon, M.S.; Self, W.H.; Courtney, D.M.; Nowak, R.M.; et al. Serial Procalcitonin Predicts Mortality in Severe Sepsis Patients: Results from the Multicenter Procalcitonin MOnitoring SEpsis (MOSES) Study. Crit. Care Med. 2017, 45, 781–789. [Google Scholar] [CrossRef] [PubMed]
- Wacker, C.; Prkno, A.; Brunkhorst, F.M.; Schlattmann, P. Procalcitonin as a diagnostic marker for sepsis: A systematic review and meta-analysis. Lancet Infect. Dis. 2013, 13, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Schuetz, P.; Beishuizen, A.; Broyles, M.; Ferrer, R.; Gavazzi, G.; Gluck, E.H.; del Castillo, J.G.; Jensen, J.-U.; Kanizsai, P.L.; Kwa, A.L.H.; et al. Procalcitonin (PCT)-guided antibiotic stewardship: An international experts consensus on optimized clinical use. Clin. Chem. Lab. Med. 2019, 57, 1308–1318. [Google Scholar] [CrossRef] [PubMed]
- de Jong, E.; van Oers, J.A.; Beishuizen, A.; Vos, P.; Vermeijden, W.J.; E Haas, L.; Loef, B.G.; Dormans, T.; van Melsen, G.C.; Kluiters, Y.C.; et al. Efficacy and safety of procalcitonin guidance in reducing the duration of antibiotic treatment in critically ill patients: A randomised, controlled, open-label trial. Lancet Infect. Dis. 2016, 16, 819–827. [Google Scholar] [CrossRef] [PubMed]
- Tong-Minh, K.; van der Does, Y.; Engelen, S.; de Jong, E.; Ramakers, C.; Gommers, D.; van Gorp, E.; Endeman, H. High procalcitonin levels associated with increased intensive care unit admission and mortality in patients with a COVID-19 infection in the emergency department. BMC Infect. Dis. 2022, 22, 165. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wu, C.; Zhang, Q.; Wu, F.; Yu, B.; Lv, J.; Li, Y.; Li, T.; Zhang, S.; Wu, C.; et al. C-Reactive Protein Level May Predict the Risk of COVID-19 Aggravation. Open Forum Infect. Dis. 2020, 7, ofaa153. [Google Scholar] [CrossRef] [PubMed]
- Ali, N. Elevated level of C-reactive protein may be an early marker to predict risk for severity of COVID-19. J. Med. Virol. 2020, 92, 2409–2411. [Google Scholar] [CrossRef] [PubMed]
- Gregoriano, C.; Heilmann, E.; Molitor, A.; Schuetz, P. Role of procalcitonin use in the management of sepsis. J. Thorac Dis. 2020, 12 (Suppl. S1), S5–S15. [Google Scholar] [CrossRef] [PubMed]
- Alnimr, A.M.; Alshahrani, M.S.; Alwarthan, S.; AlQahtani, S.Y.; Hassan, A.A.; BuMurah, N.N.; Alhajiri, S.; Bukharie, H. Bacterial and fungal coinfection in critically ill COVID-19 cases and predictive role of procalcitonin during the first wave at an academic health center. J. Epidemiol. Glob. Health 2022, 12, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Galloway, A. Non-probability sampling. In Encyclopedia of Social Measurement; Elsevier: Amsterdam, The Netherlands, 2005; pp. 859–864. [Google Scholar] [CrossRef]
- Harrell, F.E., Jr.; Dupont, C. Hmisc: Harrell Miscellaneous [Internet], Version 5.1-1; Published 12 September 2023. Available online: https://hbiostat.org/R/Hmisc/ (accessed on 20 December 2023).
- Harrell, F.E., Jr. Regression Modeling Strategies: 15 Regression Models for Continuous Y and Case Studies in Ordinal Regression. HBI Biostatistics. 2023. Available online: https://hbiostat.org/rmsc/cony.html#ordinal-regression-models-for-continuous-y (accessed on 25 February 2024).
- Bürkner, P.C.; Gabry, J.; Weber, S.; Johnson, A.; Modrak, M.; Badr, H.S.; Weber, F.; Ben-Shachar, M.S.; Rabel, H.; Mills, S.C.; et al. CRAN-Package brms: Bayesian Regression Models Using ‘Stan’. The Comprehensive R Archive Network (CRAN). 25 September 2023. Available online: https://cran.r-project.org/web/packages/brms/index.html (accessed on 25 February 2024).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. Available online: https://ggplot2.tidyverse.org (accessed on 12 January 2024).



| Characteristic | Normal Procalcitonin (n = 162) | Elevated Procalcitonin (n = 78) | p-Value |
|---|---|---|---|
| Age, years, median (IQR) | 57.5 (46.7–70.0) | 64.0 (50.3–72.8) | 0.269 |
| Male, n (%) | 92 (56.8) | 47 (60.3) | 0.711 |
| Weight, kg, median (IQR) | 85.5 (72.5–96.0) | 76.0 (64.0–93.8) | 0.069 |
| SBP, mmHg, mean (SD) | 133.0 (117.8–146.0) | 128.5 (117.0–140.5) | 0.133 |
| DBP, mmHg, mean (SD) | 71.54 (63.75–79.25) | 70.27 (70.0–77.5) | 0.465 |
| BUN, mg/dL, median (IQR) | 4.8 (3.4–7.2) | 7.6 (4.8–13.3) | <0.001 |
| SCr, mmol/L, median (IQR) | 83.0 (67.0–102.0) | 105.5 (76.7–206.5) | <0.001 |
| Lactate, mmol/L, median (IQR) | 1.4 (1.0–1.8) | 1.5 (1.0–1.9) | 0.294 |
| WBC, ×109/L, median (IQR) | 5.6 (4.1–7.3) | 7.8 (5.1–11.4) | <0.001 |
| Neutrophils in %, median (IQR) | 67.4 (57.7–77.0) | 76.0 (64.6–83.9) | <0.001 |
| Lymphocyte in %, median (IQR) | 23.1 (15.50–30.2) | 16.9 (8.1–22.2) | <0.001 |
| CURB-65, median (IQR) | 1.0 (0–1) | 1.0 (0.0–2.0) | 0.009 |
| Comorbidities | |||
| Hypertension, n (%) | 79 (48.8) | 46 (59.0) | 0.179 |
| Diabetes, n (%) | 69 (42.6) | 43 (55.1) | 0.092 |
| Heart disease, n (%) | 34 (21.0) | 23 (29.5) | 0.198 |
| Kidney disease, n (%) | 26 (16.0) | 30 (38.5) | <0.001 |
| Liver disease, n (%) | 10 (6.2) | 1 (1.3) | 0.171 |
| Lung disease, n (%) | 18 (11.1) | 6 (7.7) | 0.550 |
| Immunocompromised status, n (%) | 12 (7.4) | 8 (10.3) | 0.618 |
| Neoplastic disease, n (%) | 1 (0.6) | 4 (5.1) | 0.070 |
| Cerebrovascular disease, n (%) | 15 (9.3) | 8 (10.3) | 0.991 |
| Microbiological Data * | |||
| Positive respiratory culture, n (%) | 3 (3.4) | 5 (6.4) | 0.225 |
| Positive blood culture, n (%) | 9 (6.6) | 8 (10.3) | 0.459 |
| Positive respiratory or blood culture, n (%) | 11 (6.8) | 13 (16.7) | 0.031 |
| Procalcitonin level, ng/mL, median (IQR) | 0.12 (0.05–0.13) | 4.04 (0.35–2.27) | <0.001 |
| Antibiotics on Admission | |||
| Azithromycin, n (%) | 140 (87.5) | 61 (78.2) | 0.096 |
| Ceftriaxone, n (%) | 133 (83.1) | 61 (78.2) | 0.459 |
| Other, n (%) † | 57 (33.3) | 31 (40.3) | 0.362 |
| Outcome | Normal Procalcitonin n = 162 | Elevated Procalcitonin n = 78 | Absolute Median Difference (95% Confidence Interval) | Estimated Median Difference (95% Credible Interval) | Odds Ratio (95% Credible Interval) |
|---|---|---|---|---|---|
| LOS, median (IQR) | 6.0 (4.0–8.0) | 7.0 (5.0–8.0) | 1.0 (−0.04 to not estimated) | 0.73 (0.25–1.21) | 2.0 (1.3–3.4) |
| Antibiotics duration, median (IQR) | 7.0 (4.0–10.0) | 8.5 (5.0–15.8) | 1.5 (−3.0 to 1.5) | 0.21 (−0.25 to 0.68) | 1.2 (0.8–1.9) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almulhim, A.S.; Alabdulwahed, M.A.; Aldoughan, F.F.; Aldayyen, A.M.; Alghamdi, F.; Alabdulqader, R.; Alnaim, N.; Alghannam, D.; Aljamaan, Y.; Almutairi, S.; et al. Evaluation of Serial Procalcitonin Levels for the Optimization of Antibiotic Use in Non-Critically Ill COVID-19 Patients. Pharmaceuticals 2024, 17, 624. https://doi.org/10.3390/ph17050624
Almulhim AS, Alabdulwahed MA, Aldoughan FF, Aldayyen AM, Alghamdi F, Alabdulqader R, Alnaim N, Alghannam D, Aljamaan Y, Almutairi S, et al. Evaluation of Serial Procalcitonin Levels for the Optimization of Antibiotic Use in Non-Critically Ill COVID-19 Patients. Pharmaceuticals. 2024; 17(5):624. https://doi.org/10.3390/ph17050624
Chicago/Turabian StyleAlmulhim, Abdulaziz S., Mohammed A. Alabdulwahed, Fatimah F. Aldoughan, Ali M. Aldayyen, Faisal Alghamdi, Rawan Alabdulqader, Norah Alnaim, Dimah Alghannam, Yasmin Aljamaan, Saleh Almutairi, and et al. 2024. "Evaluation of Serial Procalcitonin Levels for the Optimization of Antibiotic Use in Non-Critically Ill COVID-19 Patients" Pharmaceuticals 17, no. 5: 624. https://doi.org/10.3390/ph17050624








