Unveiling Novel Kunitz- and Waprin-Type Toxins in the Micrurus mipartitus Coral Snake Venom Gland: An In Silico Transcriptome Analysis
Abstract
:1. Introduction
2. Results
2.1. Transcriptomic Results
2.1.1. General Transcriptomic Assembly Analysis and Annotation
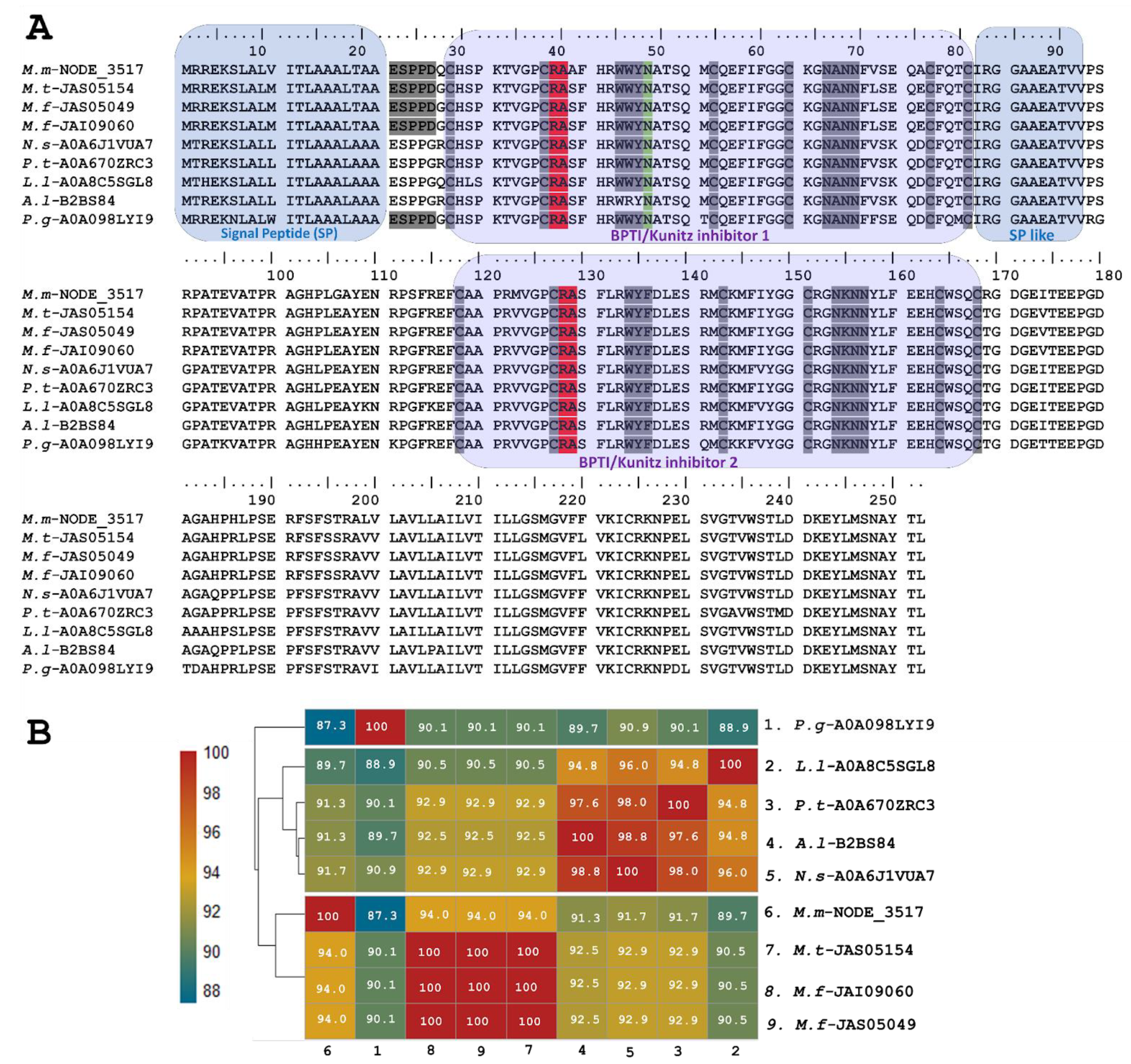
2.1.2. Transcriptomic Analysis Revealed Novel Sequences of Kunitz-Type Inhibitors (KSPI)
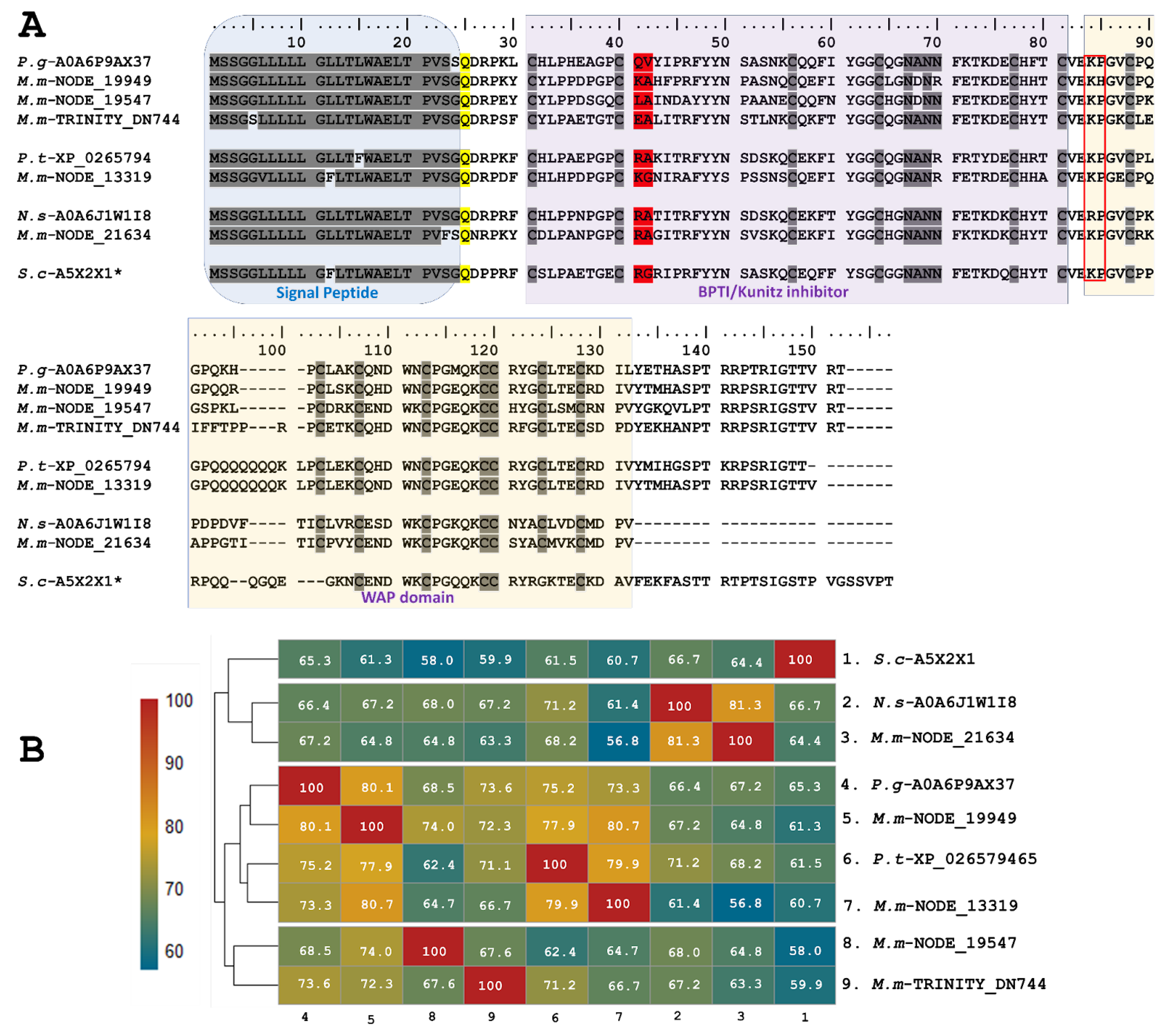
2.1.3. Single and Double Waprin Domains in the M. mipartitus Transcriptome
2.1.4. Multidomain Putative Annotated as Kunitz by ToxCodAn Shows Similarity to SPINT1 Human Gene
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Biological Samples and RNA Extraction from Venom Gland Tissue
5.2. Transcriptome Sequencing and Quality Analysis
5.3. De Novo Assembly and Quality Control Analysis
5.4. Functional Annotation of Transcriptome
5.5. Manual Sequence Curation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Drofenik, S.; Leonardi, A.; Žužek, M.C.; Frangež, R.; Križaj, I. The first Kunitz-type proteins from a viperid venom that potentiate neuromuscular transmission. Toxicon 2020, 187, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Droctové, L.; Ciolek, J.; Mendre, C.; Chorfa, A.; Huerta, P.; Carvalho, C.; Gouin, C.; Lancien, M.; StanajicPetrovic, G.; Braco, L.; et al. A new Kunitz-type snake toxin family associated with an original mode of interaction with the vasopressin 2 receptor. Br. J. Pharmacol. 2022, 179, 3470–3481. [Google Scholar] [CrossRef] [PubMed]
- Tasoulis, T.; Isbister, G.K. A Review and Database of Snake Venom Proteomes. Toxins 2017, 9, 290. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Doley, R.; Pahari, S.; Reza, M.A.; Mackessy, S.P.; Kini, R.M. The gene structure and evolution of ku-wap-fusin (Kunitz Waprin Fusion Protein), a novel evolutionary intermediate of the Kunitz Serine Protease Inhibitors and Waprins from Sistrurus catenatus (Massasauga Rattlesnake) Venom Glands. Open Evol. J. 2010, 4, 31–41. [Google Scholar]
- Mishra, M. Evolutionary aspects of the structural convergence and functional diversification of Kunitz-Domain Inhibitors. J. Mol. Evol. 2020, 88, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Kodama, R.T.; Kuniyoshi, A.K.; da Silva, C.C.; Cajado-Carvalho, D.; Duzzi, B.; Mariano, D.C.; Pimenta, D.C.; Borges, R.; da Silva, W.D.; Portaro, F.C. A Kunitz-type peptide from Dendroaspis polylepis venom as a simultaneous inhibitor of serine and cysteine proteases. J. Venom. Anim. Toxins Incl. Trop. Dis. 2020, 26, e20200037. [Google Scholar] [CrossRef] [PubMed]
- Zupunski, V.; Kordis, D.; Gubensek, F. Adaptive evolution in the snake venom Kunitz/BPTI protein family. FEBS Lett. 2003, 547, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Aird, S.D.; da Silva, N.J.; Qiu, L.; Villar-Briones, A.; Saddi, V.A.; Pires de Campos Telles, M.; Grau, M.L.; Mikheyev, A.S. Coralsnake Venomics: Analyses of Venom Gland transcriptomes and proteomes of six Brazilian Taxa. Toxins 2017, 9, 187. [Google Scholar] [CrossRef] [PubMed]
- Vincent, J.P.; Lazdunski, M. The interaction between alpha chymotrypsin and pancreatic trypsin inhibitor (Kunitz inhibitor), Kinetic and thermodynamic properties. Eur. J. Biochem. 1973, 38, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Strydom, D.J. Snake venom toxins. Purification and properties of low-molecular-weight polypeptides of Dendroaspis polylepis polylepis (black mamba) venom. Eur. J. Biochem. 1976, 69, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Sigle, R.; Hackett, M.; Aird, S.D. Primary structures of four trypsin inhibitor E homologs from venom of Dendroaspis angusticeps: Structure-function comparisons with other dendrotoxin homologs. Toxicon 2002, 40, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Harvey, A.L. Twenty years of dendrotoxins. Toxicon 2001, 39, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Jackson, T.N.; Sunagar, K.; Undheim, E.A.; Koludarov, I.; Chan, A.H.; Sanders, K.; Ali, S.A.; Hendrikx, I.; Dunstan, N.; Fry, B.G. Venom down under: Dynamic evolution of Australian elapid snake toxins. Toxins 2013, 18, 2621–2655. [Google Scholar] [CrossRef] [PubMed]
- Junqueira-de-Azevedo, I.L.; Campos, P.F.; Ching, A.T.; Mackessy, S.P. Colubrid Venom Composition: An -Omics Perspective. Toxins 2016, 8, 230. [Google Scholar] [CrossRef] [PubMed]
- St Pierre, L.; Earl, S.T.; Filippovich, I.; Sorokina, N.; Masci, P.P.; De Jersey, J.; Lavin, M.F. Common evolution of waprin and kunitz-like toxin families in Australian venomous snakes. Cell. Mol. Life Sci. 2008, 65, 4039–4054. [Google Scholar] [CrossRef] [PubMed]
- Oguiura, N.; Sanchez, L.; Duarte, P.V.; Sulca-López, M.A.; Machini, M.T. Past, present, and future of naturally occurring antimicrobials related to snake venoms. Animals 2023, 13, 744. [Google Scholar] [CrossRef] [PubMed]
- Nair, D.G.; Fry, B.G.; Alewood, P.; Kumar, P.P.; Kini, R.M. Antimicrobial activity of omwaprin, a new member of the waprin family of snake venom proteins. Biochem. J. 2007, 402, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Campos, P.F.; Andrade-Silva, D.; Zelanis, A.; Paes Leme, A.F.; Rocha, M.M.; Menezes, M.C.; Serrano, S.M.; Junqueira-de-Azevedo, I.d.L. Trends in the evolution of snake toxins underscored by an integrative omics approach to profile the venom of the Colubrid Phalotris mertensi. Genome Biol. Evol. 2016, 8, 2266–2287. [Google Scholar] [CrossRef] [PubMed]
- Silvane, L.; Celias, D.P.; Romagnoli, P.A.; Maletto, B.A.; Sanchez Vallecillo, M.F.; Chiapello, L.S.; Palma, S.D.; Allemandi, D.A.; Sanabria, R.E.; Pruzzo, C.I.; et al. A Vaccine based on Kunitz-Type molecule confers protection against fasciola hepatica challenge by inducing IFN-γ and antibody immune responses through IL-17A production. Front. Immunol. 2020, 11, 2087. [Google Scholar] [CrossRef] [PubMed]
- Sintsova, O.; Gladkikh, I.; Chausova, V.; Monastyrnaya, M.; Anastyuk, S.; Chernikov, O.; Yurchenko, E.; Aminin, D.; Isaeva, M.; Leychenko, E.; et al. Peptide fingerprinting of the sea anemone Heteractis magnifica mucus revealed neurotoxins, Kunitz-type proteinase inhibitors and a new β-defensin α-amylase inhibitor. J. Proteom. 2018, 173, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Ranasinghe, S.L.; Duke, M.; Harvie, M.; McManus, D.P. Kunitz-type protease inhibitor as a vaccine candidate against schistosomiasis mansoni. Int. J. Infect. Dis. 2018, 66, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Wang, Y.; Wei, L.; Ye, H.; Liu, H.; Wang, L.; Liu, R.; Li, D.; Lai, R. Snake venom-like waprin from the frog of Ceratophrys calcarata contains antimicrobial function. Gene 2013, 514, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Wüster, W. Assembling an arsenal: Origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences. Mol. Biol. Evol. 2004, 21, 870–883. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.G.; Scheib, H.; van der Weerd, L.; Young, B.; McNaughtan, J.; Ramjan, S.F.; Vidal, N.; Poelmann, R.E.; Norman, J.A. Evolution of an arsenal: Structural and functional diversification of the venom system in the advanced snakes (Caenophidia). Mol. Cell. Proteom. 2008, 7, 215–246. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, A.L.; Viegas, M.F.; da Silva, S.L.; Soares, A.M.; Ramos, M.J.; Fernandes, P.A. The chemistry of snake venom and its medicinal potential. Nat. Rev. Chem. 2022, 6, 451–469. [Google Scholar] [CrossRef] [PubMed]
- Lomonte, B.; Calvete, J.J.; Fernández, J.; Pla, D.; Rey-Suárez, P.; Sanz, L.; Gutiérrez, J.M.; Sasa, M. Venomic analyses of coralsnakes. In The Biology of the Coralsnakes; Eagle Mountain Publishing: Eagle Mountain, UT, USA, 2021; pp. 485–518. [Google Scholar]
- Sanz, L.; Quesada-Bernat, S.; Ramos, T.; Casais-E-Silva, L.L.; Corrêa-Netto, C.; Silva-Haad, J.J.; Sasa, M.; Lomonte, B.; Calvete, J.J. New insights into the phylogeographic distribution of the 3FTx/PLA2 venom dichotomy across genus Micrurus in South America. J. Proteom. 2019, 200, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Lomonte, B.; Sasa, M.; Rey-Suárez, P.; Bryan, W.; Gutiérrez, J.M. Venom of the Coral Snake Micrurus clarki: Proteomic profile, toxicity, immunological cross-neutralization, and characterization of a Three-Finger Toxin. Toxins 2016, 8, 138. [Google Scholar] [CrossRef] [PubMed]
- Mena, G.; Chaves-Araya, S.; Chacón, J.; Török, E.; Török, F.; Bonilla, F.; Sasa, M.; Gutiérrez, J.M.; Lomonte, B.; Fernández, J. Proteomic and toxicological analysis of the venom of Micrurus yatesi and its neutralization by an antivenom. Toxicon 2022, 13, 100097. [Google Scholar] [CrossRef] [PubMed]
- Sanz, L.; Pla, D.; Pérez, A.; Rodríguez, Y.; Zavaleta, A.; Salas, M.; Lomonte, B.; Calvete, J.J. Venomic analysis of the poorly studied desert Coral Snake, Micrurus tschudii tschudii, supports the 3FTx/PLA2 dichotomy across Micrurus Venoms. Toxins 2016, 8, 178. [Google Scholar] [CrossRef] [PubMed]
- Rey-Suárez, P.; Núñez, V.; Gutiérrez, J.M.; Lomonte, B. Proteomic and biological characterization of the venom of the redtail coral snake, Micrurus mipartitus (Elapidae), from Colombia and Costa Rica. J. Proteom. 2011, 75, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Corrêa-Netto, C.; Junqueira-de-Azevedo, I.d.L.; Silva, D.A.; Ho, P.L.; Leitão-de-Araújo, M.; Alves, M.L.; Sanz, L.; Foguel, D.; Zingali, R.B.; Calvete, J.J. Snake venomics and venom gland transcriptomic analysis of Brazilian coral snakes, Micrurus altirostris and M. corallinus. J. Proteom. 2011, 74, 1795–1809. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Vargas, A.; Franco-Vásquez, A.M.; Bolívar-Barbosa, J.A.; Vega, N.; Reyes-Montaño, E.; Arreguín-Espinosa, R.; Carbajal-Saucedo, A.; Angarita-Sierra, T.; Ruiz-Gómez, F. Unveiling the venom composition of the Colombian Coral Snakes Micrurus helleri, M. medemi, and M. sangilensis. Toxins 2023, 15, 622. [Google Scholar] [CrossRef] [PubMed]
- Bénard-Valle, M.; Neri-Castro, E.; Yañez-Mendoza, M.F.; Lomonte, B.; Olvera, A.; Zamudio, F.; Restano-Cassulini, R.; Possani, L.D.; Jiménez-Ferrer, E.; Alagón, A. Functional, proteomic and transcriptomic characterization of the venom from Micrurus browni browni: Identification of the first lethal multimeric neurotoxin in coral snake venom. J. Proteom. 2020, 225, 103863. [Google Scholar] [CrossRef] [PubMed]
- Olamendi-Portugal, T.; Batista, C.V.; Pedraza-Escalona, M.; Restano-Cassulini, R.; Zamudio, F.Z.; Benard-Valle, M.; de Roodt, A.R.; Possani, L.D. New insights into the proteomic characterization of the coral snake Micrurus pyrrhocryptus venom. Toxicon 2018, 153, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Rey-Suárez, P.; Núñez, V.; Fernández, J.; Lomonte, B. Integrative characterization of the venom of the coral snake Micrurus dumerilii (Elapidae) from Colombia: Proteome, toxicity, and cross-neutralization by antivenom. J. Proteom. 2016, 136, 262–273. [Google Scholar] [CrossRef] [PubMed]
- Fernández, J.; Vargas-Vargas, N.; Pla, D.; Sasa, M.; Rey-Suárez, P.; Sanz, L.; Gutiérrez, J.M.; Calvete, J.J.; Lomonte, B. Snake venomics of Micrurus alleni and Micrurus mosquitensis from the Caribbean region of Costa Rica reveals two divergent compositional patterns in New World elapids. Toxicon 2015, 107, 217–233. [Google Scholar] [CrossRef] [PubMed]
- Lippa, E.; Török, F.; Gómez, A.; Corrales, G.; Chacón, D.; Sasa, M.; Gutiérrez, J.M.; Lomonte, B.; Fernández, J. First look into the venom of Roatan Island’s critically endangered coral snake Micrurus ruatanus: Proteomic characterization, toxicity, immunorecognition and neutralization by an antivenom. J. Proteom. 2019, 198, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Fernández, J.; Alape-Girón, A.; Angulo, Y.; Sanz, L.; Gutiérrez, J.M.; Calvete, J.J.; Lomonte, B. Venomic and antivenomic analyses of the Central American coral snake, Micrurus nigrocinctus (Elapidae). J. Proteome Res. 2011, 10, 1816–1827. [Google Scholar] [CrossRef] [PubMed]
- Aird, S.D.; da Silva, N.J. Venom yields of Brazilian coral snakes. In Advances in Coralsnake Biology with an Emphasis on South America; Eagle Mountain Publishing: Eagle Mountain, UT, USA, 2021; ISBN 978-0972015462. [Google Scholar]
- Margres, M.J.; Aronow, K.; Loyacano, J.; Rokyta, D.R. The venom-gland transcriptome of the eastern coral snake (Micrurus fulvius) reveals high venom complexity in the intragenomic evolution of venoms. BMC Genom. 2013, 14, 531. [Google Scholar] [CrossRef] [PubMed]
- Bénard-Valle, M.; Neri-Castro, E.; Elizalde-Morales, N.; Olvera-Rodríguez, A.; Strickland, J.; Acosta, G.; Alagón, A. Protein composition and biochemical characterization of venom from Sonoran Coral Snakes (Micruroides euryxanthus). Biochimie 2021, 182, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Altamirano, J.A.; Salazar-Valenzuela, D.; Medina-Villamizar, E.J.; Quirola, D.R.; Patel, K.; Vaiyapuri, S.; Lomonte, B.; Almeida, J.R. First Insights into the Venom Composition of Two Ecuadorian Coral Snakes. Int. J. Mol. Sci. 2022, 23, 14686. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Župunski, V.; Kordiš, D. Strong and widespread action of site-specific positive selection in the snake venom Kunitz/BPTI protein family. Sci. Rep. 2016, 6, 37054. [Google Scholar] [CrossRef] [PubMed]
- Munawar, A.; Ali, S.A.; Akrem, A.; Betzel, C. Snake Venom Peptides: Tools of Biodiscovery. Toxins 2018, 10, 474. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, H. Snake Venoms. In Snake Venom Protease Inhibitors: Enhanced Identification, Expanding Biological Function, and Promising Future; Springer: Dordrecht, The Netherlands, 2015. [Google Scholar] [CrossRef]
- Slagboom, J.; Kool, J.; Harrison, R.A.; Casewell, N.R. Haemotoxic snake venoms: Their functional activity, impact on snakebite victims and pharmaceutical promise. Br. J. Haematol. 2017, 177, 947–959. [Google Scholar] [CrossRef]
- Earl, S.T.; Richards, R.; Johnson, L.A.; Flight, S.; Anderson, S.; Liao, A.; de Jersey, J.; Masci, P.P.; Lavin, M.F. Identification and characterisation of Kunitz-type plasma kallikrein inhibitors unique to Oxyuranus sp. snake venoms. Biochimie 2012, 94, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Fry, B. Venomous Reptiles and Their Toxins: Evolution, Pathophysiology, and Biodiscovery; Oxford University Press: New York, NY, USA, 2015; ISBN 9780199309399. [Google Scholar]
- Rokyta, D.R.; Margres, M.J.; Calvin, K. Post-transcriptional Mechanisms Contribute Little to Phenotypic Variation in Snake Venoms. G3 2015, 5, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Ritonja, A.; Turk, V.; Gubensek, F. Serine proteinase inhibitors from Vipera ammodytes venom. Isolation and kinetic studies. Eur. J. Biochem. 1983, 133, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Yang, H.; Yu, H.; Gao, W.; Lai, R.; Liu, J.; Liang, X. A novel serine protease inhibitor from Bungarus fasciatus venom. Peptides 2008, 29, 369–374. [Google Scholar] [CrossRef]
- Thakur, R.; Mukherjee, A.K. Pathophysiological significance and therapeutic applications of snake venom protease inhibitors. Toxicon 2017, 131, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Vivas, J.; Ibarra, C.; Salazar, A.M.; Neves-Ferreira, A.G.; Sánchez, E.E.; Perales, J.; Rodríguez-Acosta, A.; Guerrero, B. Purification and characterization of tenerplasminin-1, a serine peptidase inhibitor with antiplasmin activity from the coral snake (Micrurus tener tener) venom. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2016, 179, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Bohlen, C.J.; Chesler, A.T.; Sharif-Naeini, R.; Medzihradszky, K.F.; Zhou, S.; King, D.; Sánchez, E.E.; Burlingame, A.L.; Basbaum, A.I.; Julius, D. A heteromeric Texas coral snake toxin targets acid-sensing ion channels to produce pain. Nature 2011, 479, 410–414. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.Y.; Liew, J.L.; Tan, N.H.; Quah, E.S.; Ismail, A.K.; Tan, C.H. Unlocking the secrets of banded coral snake (Calliophis intestinalis, Malaysia): A venom with proteome novelty, low toxicity and distinct antigenicity. J. Proteom. 2019, 192, 246–257. [Google Scholar] [CrossRef] [PubMed]
- Lomonte, B.; Rey-Suárez, P.; Fernández, J.; Sasa, M.; Pla, D.; Vargas, N.; Bénard-Valle, M.; Sanz, L.; Corrêa-Netto, C.; Núñez, V.; et al. Venoms of Micrurus coral snakes: Evolutionary trends in compositional patterns emerging from proteomic analyses. Toxicon 2016, 122, 7–25. [Google Scholar] [CrossRef] [PubMed]
- Jowers, M.J.; Smart, U.; Sánchez-Ramírez, S.; Murphy, J.C.; Gómez, A.; Bosque, R.J.; Sarker, G.C.; Noonan, B.P.; Faria, J.F.; Harris, D.J.; et al. Unveiling underestimated species diversity within the Central American Coralsnake, a medically important complex of venomous taxa. Sci. Rep. 2023, 13, 11674. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.M.; Wong, H.Y.; Desai, M.; Moochhala, S.; Kuchel, P.W.; Kini, R.M. Identification of a novel family of proteins in snake venoms. Purification and structural characterization of nawaprin from Naja nigricollis snake venom. J. Biol. Chem. 2003, 278, 40097–40104. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Nagaike, K.; Takeda, N.; Itoh, H.; Kohama, K.; Fukushima, T.; Miyata, S.; Uchiyama, S.; Uchinokura, S.; Shimomura, T.; et al. Hepatocyte growth factor activator inhibitor type 1 (HAI-1) is required for branching morphogenesis in the chorioallantoic placenta. Mol. Cell. Biol. 2005, 25, 5687–5698. [Google Scholar] [CrossRef]
- Kataoka, H.; Miyata, S.; Uchinokura, S.; Itoh, H. Roles of hepatocyte growth factor (HGF) activator and HGF activator inhibitor in the pericellular activation of HGF/scatter factor. Cancer Metastasis Rev. 2003, 22, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Urra, F.A.; Araya-Maturana, R. Targeting Metastasis with Snake Toxins: Molecular Mechanisms. Toxins 2017, 9, 390. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Morita, T. Snake venom components affecting blood coagulation and the vascular system: Structural similarities and marked diversity. Curr. Pharm. Des. 2007, 13, 28726–28786. [Google Scholar] [CrossRef] [PubMed]
- Pla, D.; Petras, D.; Saviola, A.J.; Modahl, C.M.; Sanz, L.; Pérez, A.; Juárez, E.; Frietze, S.; Dorrestein, P.C.; Mackessy, S.P.; et al. Transcriptomics-guided bottom-up and top-down venomics of neonate and adult specimens of the arboreal rear-fanged Brown Treesnake, Boiga irregularis, from Guam. J. Proteom. 2018, 74, 716–784. [Google Scholar] [CrossRef] [PubMed]
- Gioia, M.; Ciaccio, C.; Calligari, P.; De Simone, G.; Sbardella, D.; Tundo, G.; Fasciglione, G.F.; Di Masi, A.; Di Pierro, D.; Bocedi, A.; et al. Role of proteolytic enzymes in the COVID-19 infection and promising therapeutic approaches. Biochem. Pharmacol. 2020, 182, 114225. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Tian, M.; Qi, W.; Wu, J.; Zheng, H.; Guo, G.; Zhang, L.; Ranasinghe, S.L.; McManus, D.P.; Zhang, W. Bioinformatic comparison of Kunitz protease inhibitors in Echinococcus granulosus sensu stricto and E. multilocularis and the genes expressed in different developmental stages of E. granulosus s.s. BMC Genom. 2021, 22, 907. [Google Scholar] [CrossRef] [PubMed]
- Bonturi, C.R.; Silva Teixeira, A.B.; Rocha, V.M.; Valente, P.F.; Oliveira, J.R.; Filho, C.M.; Batista, I.F.C.; Oliva, M.L.V. Plant Kunitz Inhibitors and their interaction with proteases: Current and potential pharmacological targets. Int. J. Mol. Sci. 2022, 23, 4742. [Google Scholar] [CrossRef] [PubMed]
- Fratini, E.; Rossi, M.N.; Spagoni, L.; Riccieri, A.; Mancini, E.; Polticelli, F.; Bologna, M.A.; Mariottini, P.; Cervelli, M. Molecular characterization of Kunitz-Type protease inhibitors from Blister Beetles (Coleoptera, Meloidae). Biomolecules 2022, 12, 988. [Google Scholar] [CrossRef] [PubMed]
- Kleiz-Ferreira, J.M.; Bernaerts, H.; Pinheiro-Junior, E.L.; Peigneur, S.; Zingali, R.B.; Tytgat, J. Pharmacological screening of venoms from five Brazilian Micrurus species on different ion channels. Int. J. Mol. Sci. 2022, 23, 7714. [Google Scholar] [CrossRef] [PubMed]
- Ciolek, J.; Reinfrank, H.; Quinton, L.; Viengchareun, S.; Stura, E.A.; Vera, L.; Sigismeau, S.; Mouillac, B.; Orcel, H.; Peigneur, S.; et al. Green mamba peptide targets type-2 vasopressin receptor against polycystic kidney disease. Proc. Natl. Acad. Sci. USA 2017, 114, 7154–7159. [Google Scholar] [CrossRef] [PubMed]
- Morjen, M.; Moslah, W.; Touihri-Baraketi, I.; Srairi-Abid, N.; Luis, J.; Marrakchi, N.; Jebali, J. Expression of the first recombinant anti-tumoral snake venom Kunitz-Type Serine Protease Inhibitor. Toxins 2022, 14, 170. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Goenaga, J.; López-Abán, J.; Protasio, A.V.; Santiago, B.V.; Del Olmo, E.; Vanegas, M.; Fernández-Soto, P.; Patarroyo, M.A.; Muro, A. Peptides derived of Kunitz-Type Serine Protease Inhibitor as potential vaccine against experimental Schistosomiasis. Front. Immunol. 2019, 10, 2498. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Rokyta, D.R.; Lemmon, A.R.; Margres, M.J.; Aronow, K. The venom-gland transcriptome of the eastern diamondback rattlesnake (Crotalus adamanteus). BMC Genom. 2012, 13, 312. [Google Scholar] [CrossRef] [PubMed]
- Rana, S.B.; Zadlock, F.J.; Zhang, Z.; Murphy, W.R.; Bentivegna, C.S. Comparison of de novo transcriptome assemblers and k-mer strategies using the killifish, Fundulus heteroclitus. PLoS ONE 2016, 11, e0153104. [Google Scholar] [CrossRef]
- Nachtigall, P.G.; Freitas-de-Sousa, L.A.; Mason, A.J.; Moura-da-Silva, A.M.; Grazziotin, F.G.; Junqueira-de-Azevedo, I.L. Differences in PLA2 constitution distinguish the venom of two endemic Brazilian mountain Lanceheads, Bothrops cotiara and Bothrops fonsecai. Toxins 2022, 14, 237. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wu, G.; Tang, J.; Luo, R.; Patterson, J.; Liu, S.; Huang, W.; He, G.; Gu, S.; Li, S.; et al. SOAPdenovo-Trans: De novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 2014, 30, 1660–1666. [Google Scholar] [CrossRef] [PubMed]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Nachtigall, P.G.; Rautsaw, R.M.; Ellsworth, S.A.; Mason, A.J.; Rokyta, D.R.; Parkinson, C.L.; Junqueira-de-Azevedo, I.L. ToxCodAn: A new toxin annotator and guide to venom gland transcriptomics. Brief. Bioinform. 2021, 22, bbab095. [Google Scholar] [CrossRef] [PubMed]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef] [PubMed]
- Posit Team. RStudio: Integrated Development Environment for R. Posit Software; PBC: Boston, MA, USA, 2024; Available online: http://www.posit.co/ (accessed on 5 March 2024).



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saldarriaga-Córdoba, M.; Clavero-León, C.; Rey-Suarez, P.; Nuñez-Rangel, V.; Avendaño-Herrera, R.; Solano-González, S.; Alzate, J.F. Unveiling Novel Kunitz- and Waprin-Type Toxins in the Micrurus mipartitus Coral Snake Venom Gland: An In Silico Transcriptome Analysis. Toxins 2024, 16, 224. https://doi.org/10.3390/toxins16050224
Saldarriaga-Córdoba M, Clavero-León C, Rey-Suarez P, Nuñez-Rangel V, Avendaño-Herrera R, Solano-González S, Alzate JF. Unveiling Novel Kunitz- and Waprin-Type Toxins in the Micrurus mipartitus Coral Snake Venom Gland: An In Silico Transcriptome Analysis. Toxins. 2024; 16(5):224. https://doi.org/10.3390/toxins16050224
Chicago/Turabian StyleSaldarriaga-Córdoba, Mónica, Claudia Clavero-León, Paola Rey-Suarez, Vitelbina Nuñez-Rangel, Ruben Avendaño-Herrera, Stefany Solano-González, and Juan F. Alzate. 2024. "Unveiling Novel Kunitz- and Waprin-Type Toxins in the Micrurus mipartitus Coral Snake Venom Gland: An In Silico Transcriptome Analysis" Toxins 16, no. 5: 224. https://doi.org/10.3390/toxins16050224





