Ectopic MYBL2-Mediated Regulation of Androglobin Gene Expression
Abstract
:1. Introduction
2. Materials and Methods
2.1. Expression Plasmid Constructs
2.2. Mammalian Cell Culture and DNA Transfection
2.3. Luciferase Reporter Gene Assays
2.4. dCas9-Mediated Interference of Transcription Factor Binding
2.5. Chromatin Immunoprecipitation (ChIP)
2.6. Protein Extraction and Immunoblotting
2.7. RNA Extraction and RT-qPCR
2.8. In Silico Analysis of Binding Sites
2.9. ChIP-Sequencing Analysis
2.10. Data Analysis
3. Results
3.1. The ADGB Promoter Is Regulated by Multiple Candidate Transcription Factors
3.2. Identification of the Main Binding Region of MYBL2 and PITX2 within the ADGB Promoter
3.3. MYBL2 Binding to the ADGB Promoter Is Impaired by dCas9-Mediated Blocking and Mutation of the Putative Binding Site
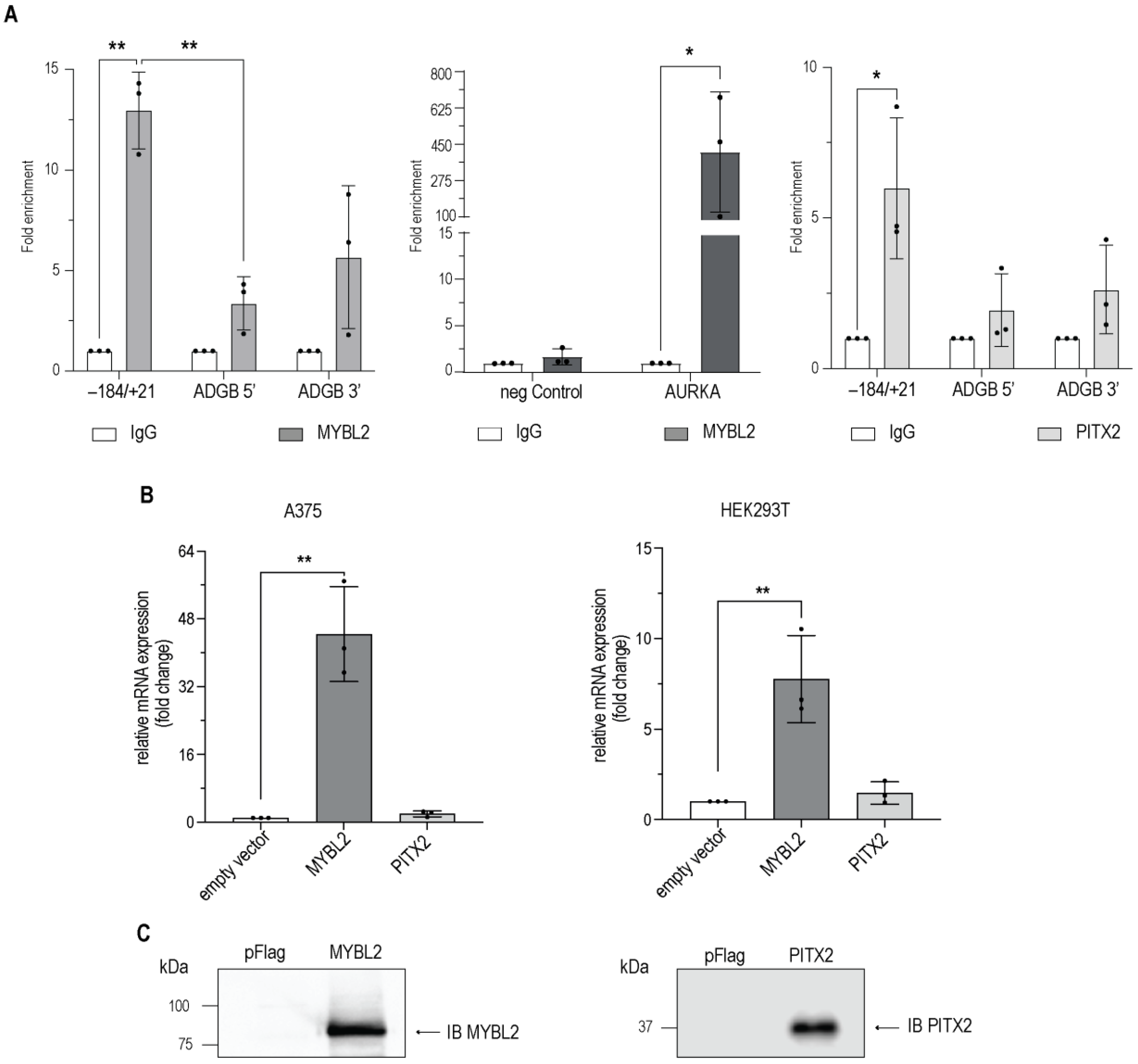
3.4. MYBL2 and PITX2 Interact with the Endogenous ADGB Promoter
4. Discussion
5. Limitations of the Study
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Burmester, T.; Hankeln, T. Function and evolution of vertebrate globins. Acta Physiol. 2014, 211, 501–514. [Google Scholar] [CrossRef]
- Keppner, A.; Maric, D.; Correia, M.; Koay, T.W.; Orlando, I.M.C.; Vinogradov, S.N.; Hoogewijs, D. Lessons from the post-genomic era: Globin diversity beyond oxygen binding and transport. Redox Biol. 2020, 37, 101687. [Google Scholar] [CrossRef]
- Bracke, A.; Hoogewijs, D.; Dewilde, S. Exploring three different expression systems for recombinant expression of globins: Escherichia coli, Pichia pastoris and Spodoptera frugiperda. Anal. Biochem. 2018, 543, 62–70. [Google Scholar] [CrossRef]
- Hoogewijs, D.; Ebner, B.; Germani, F.; Hoffmann, F.G.; Fabrizius, A.; Moens, L.; Burmester, T.; Dewilde, S.; Storz, J.F.; Vinogradov, S.N.; et al. Androglobin: A chimeric globin in metazoans that is preferentially expressed in Mammalian testes. Mol. Biol. Evol. 2012, 29, 1105–1114. [Google Scholar] [CrossRef]
- Reeder, B.J.; Deganutti, G.; Ukeria, J.; Atanasioa, S.; Svistunenko, D.A.; Ronchettia, C.; Mobarec, J.C.; Welbourn, E.; Asajua, J.; Vos, M.H.; et al. The circularly permuted globin domain of Androglobin exhibits atypical heme stabilization and nitric oxide interaction. Chem. Sci. 2024, in press. [Google Scholar] [CrossRef]
- Keppner, A.; Correia, M.; Santambrogio, S.; Koay, T.W.; Maric, D.; Osterhof, C.; Winter, D.V.; Clerc, A.; Stumpe, M.; Chalmel, F.; et al. Androglobin, a chimeric mammalian globin, is required for male fertility. eLife 2022, 11, e72374. [Google Scholar] [CrossRef]
- Qu, R.; Zhang, Z.; Wu, L.; Li, Q.; Mu, J.; Zhao, L.; Yan, Z.; Wang, W.; Zeng, Y.; Liu, R.; et al. ADGB variants cause asthenozoospermia and male infertility. Hum. Genet. 2023, 142, 735–748. [Google Scholar] [CrossRef]
- Goa, Y.; Liu, L.; Tian, S.; Liu, C.; Lv, M.; Wu, H.; Tang, D.; Song, B.; Shen, Q.; Xu, Y.; et al. Whole-exome sequencing identifies ADGB as a novel causative gene for male infertility in humans: From motility to fertilization. Andrology 2024, 1–12. [Google Scholar] [CrossRef]
- Koay, T.W.; Osterhof, C.; Orlando, I.M.C.; Keppner, A.; Andre, D.; Yousefian, S.; Alonso, M.S.; Correia, M.; Markworth, R.; Schodel, J.; et al. Androglobin gene expression patterns and FOXJ1-dependent regulation indicate its functional association with ciliogenesis. J. Biol. Chem. 2021, 296, 100291. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Ng, C.P.; Habacher, H.; Roy, S. Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nat. Genet. 2008, 40, 1445–1453. [Google Scholar] [CrossRef] [PubMed]
- Seiler, C.Y.; Park, J.G.; Sharma, A.; Hunter, P.; Surapaneni, P.; Sedillo, C.; Field, J.; Algar, R.; Price, A.; Steel, J.; et al. DNASU plasmid and PSI:Biology-Materials repositories: Resources to accelerate biological research. Nucleic Acids Res. 2014, 42, D1253–D1260. [Google Scholar] [CrossRef] [PubMed]
- Jordan, M.; Schallhorn, A.; Wurm, F.M. Transfecting mammalian cells: Optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 1996, 24, 596–601. [Google Scholar] [CrossRef]
- Karaghiannis, V.; Maric, D.; Garrec, C.; Maaziz, N.; Buffet, A.; Schmitt, L.; Antunes, V.; Airaud, F.; Aral, B.; Le Roy, A.; et al. Comprehensive in silico and functional studies for classification of EPAS1/HIF2A genetic variants identified in patients with erythrocytosis. Haematologica 2023, 108, 1652–1666. [Google Scholar] [CrossRef] [PubMed]
- Randi, E.B.; Vervaet, B.; Tsachaki, M.; Porto, E.; Vermeylen, S.; Lindenmeyer, M.T.; Thuy, L.T.T.; Cohen, C.D.; Devuyst, O.; Kistler, A.D.; et al. The Antioxidative Role of Cytoglobin in Podocytes: Implications for a Role in Chronic Kidney Disease. Antioxid. Redox Signal 2020, 32, 1155–1171. [Google Scholar] [CrossRef]
- De Backer, J.; Maric, D.; Zuhra, K.; Bogaerts, A.; Szabo, C.; Vanden Berghe, W.; Hoogewijs, D. Cytoglobin Silencing Promotes Melanoma Malignancy but Sensitizes for Ferroptosis and Pyroptosis Therapy Response. Antioxidants 2022, 11, 1548. [Google Scholar] [CrossRef]
- Koay, T.W.; Schödel, J.; Hoogewijs, D. CRISPR Activator Approaches to Study Endogenous Androglobin Gene Regulation. Methods Mol. Biol. 2023, 2648, 167–185. [Google Scholar]
- Fonseca Costa, S.S.; Ripperger, J.A. Chromatin Immunoprecipitation (ChIP) from Mouse Liver Nuclei. Methods Mol. Biol. 2021, 2130, 115–125. [Google Scholar] [PubMed]
- De Backer, J.; Maric, D.; Bosman, M.; Dewilde, S.; Hoogewijs, D. A reliable set of reference genes to normalize oxygen-dependent cytoglobin gene expression levels in melanoma. Sci. Rep. 2021, 11, 10879. [Google Scholar] [CrossRef]
- Storti, F.; Santambrogio, S.; Crowther, L.M.; Otto, T.; Abreu-Rodriguez, I.; Kaufmann, M.; Hu, C.J.; Dame, C.; Fandrey, J.; Wenger, R.H.; et al. A novel distal upstream hypoxia response element regulating oxygen-dependent erythropoietin gene expression. Haematologica 2014, 99, e45–e48. [Google Scholar] [CrossRef]
- Schörg, A.; Santambrogio, S.; Platt, J.L.; Schodel, J.; Lindenmeyer, M.T.; Cohen, C.D.; Schrodter, K.; Mole, D.R.; Wenger, R.H.; Hoogewijs, D. Destruction of a distal hypoxia response element abolishes trans-activation of the PAG1 gene mediated by HIF-independent chromatin looping. Nucleic Acids Res. 2015, 43, 5810–5823. [Google Scholar] [CrossRef]
- Orlando, I.M.C.; Lafleur, V.N.; Storti, F.; Spielmann, P.; Crowther, L.; Santambrogio, S.; Schodel, J.; Hoogewijs, D.; Mole, D.R.; Wenger, R.H. Distal and proximal hypoxia response elements cooperate to regulate organ-specific erythropoietin gene expression. Haematologica 2020, 105, 2774–2784. [Google Scholar] [CrossRef]
- Kel, A.E.; Gossling, E.; Reuter, I.; Cheremushkin, E.; Kel-Margoulis, O.V.; Wingender, E. MATCH (TM): A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003, 31, 3576–3579. [Google Scholar] [CrossRef]
- Matys, V.; Kel-Margoulis, O.V.; Fricke, E.; Liebich, I.; Land, S.; Barre-Dirrie, A.; Reuter, I.; Chekmenev, D.; Krull, M.; Hornischer, K.; et al. TRANSFAC (R) and its module TRANSCompel (R): Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006, 34, D108–D110. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdottir, H.; Turner, D.; Mesirov, J.P. igv.js: An embeddable JavaScript implementation of the Integrative Genomics Viewer (IGV). Bioinformatics 2023, 39, btac830. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef]
- Müller, G.A.; Quaas, M.; Schumann, M.; Krause, E.; Padi, M.; Fischer, M.; Litovchick, L.; DeCaprio, J.A.; Engeland, K. The CHR promoter element controls cell cycle-dependent gene transcription and binds the DREAM and MMB complexes. Nucleic Acids Res. 2012, 40, 1561–1578. [Google Scholar] [CrossRef] [PubMed]
- Müller, G.A.; Engeland, K. The central role of CDE/CHR promoter elements in the regulation of cell cycle-dependent gene transcription. FEBS J. 2010, 277, 877–893. [Google Scholar] [CrossRef]
- Musa, J.; Aynaud, M.M.; Mirabeau, O.; Delattre, O.; Grunewald, T.G. MYBL2 (B-Myb): A central regulator of cell proliferation, cell survival and differentiation involved in tumorigenesis. Cell Death Dis. 2017, 8, e2895. [Google Scholar] [CrossRef]
- Sadasivam, S.; Duan, S.; DeCaprio, J.A. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes. Dev. 2012, 26, 474–489. [Google Scholar] [CrossRef]
- Uhlen, M.; Karlsson, M.J.; Zhong, W.; Tebani, A.; Pou, C.; Mikes, J.; Lakshmikanth, T.; Forsstrom, B.; Edfors, F.; Odeberg, J.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Logan, M.; Pagan-Westphal, S.M.; Smith, D.M.; Paganessi, L.; Tabin, C.J. The transcription factor Pitx2 mediates situs-specific morphogenesis in response to left-right asymmetric signals. Cell 1998, 94, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Reiter, J.F.; Leroux, M.R. Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 2017, 18, 533–547. [Google Scholar] [CrossRef]
- Bellchambers, H.M.; Phatak, A.R.; Nenni, M.J.; Padua, M.B.; Gao, H.; Liu, Y.; Ware, S.M. Single cell RNA analysis of the left-right organizer transcriptome reveals potential novel heterotaxy genes. Sci. Rep. 2023, 13, 10688. [Google Scholar] [CrossRef] [PubMed]
- Padua, M.B.; Helm, B.M.; Wells, J.R.; Smith, A.M.; Bellchambers, H.M.; Sridhar, A.; Ware, S.M. Congenital heart defects caused by FOXJ1. Hum. Mol. Genet. 2023, 32, 2335–2346. [Google Scholar] [CrossRef] [PubMed]
- Rochon, E.R.; Xue, J.; Mohammed, M.S.; Smith, C.; Hay-Schmidt, A.; DeMartino, A.W.; Clark, A.; Xu, Q.; Lo, C.W.; Tsang, M.; et al. Cytoglobin regulates NO-dependent cilia motility and organ laterality during development. Nat. Commun. 2023, 14, 8333. [Google Scholar] [CrossRef] [PubMed]
- Lam, E.W.; Robinson, C.; Watson, R.J. Characterization and cell cycle-regulated expression of mouse B-myb. Oncogene 1992, 7, 1885–1890. [Google Scholar] [PubMed]
- Litovchick, L.; Sadasivam, S.; Florens, L.; Zhu, X.; Swanson, S.K.; Velmurugan, S.; Chen, R.; Washburn, M.P.; Liu, X.S.; DeCaprio, J.A. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol. Cell 2007, 26, 539–551. [Google Scholar] [CrossRef] [PubMed]
- Sadasivam, S.; DeCaprio, J.A. The DREAM complex: Master coordinator of cell cycle-dependent gene expression. Nat. Rev. Cancer 2013, 13, 585–595. [Google Scholar] [CrossRef]
- Tucker, R.W.; Pardee, A.B.; Fujiwara, K. Centriole Ciliation Is Related to Quiescence and DNA-Synthesis in 3t3-Cells. Cell 1979, 17, 527–535. [Google Scholar] [CrossRef]
- Pugacheva, E.N.; Jablonski, S.A.; Hartman, T.R.; Henske, E.P.; Golemis, E.A. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell 2007, 129, 1351–1363. [Google Scholar] [CrossRef] [PubMed]
- Engeland, K. Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ. 2018, 25, 114–132. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Patestos, N.P.; Maekawa, T.; Ishii, S. B-myb is required for inner cell mass formation at an early stage of development. J. Biol. Chem. 1999, 274, 28067–28070. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.J.; Ma’ayan, A.; Lieu, Y.K.; John, P.; Reddy, M.V.R.; Chen, E.Y.; Duan, Q.N.; Snoeck, H.W.; Reddy, E.P. B-myb is an essential regulator of hematopoietic stem cell and myeloid progenitor cell development. Proc. Natl. Acad. Sci. USA 2014, 111, 3122–3127. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Karlsson, M.J.; Zhong, W.; Tebani, A.; Pou, C.; Mikes, J.; Lakshmikanth, T.; Forsstrom, B.; Edfors, F.; Odeberg, J.; et al. A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science 2019, 366, eaax9198. [Google Scholar] [CrossRef]
- Ward, C.; Volpe, G.; Cauchy, P.; Ptasinska, A.; Almaghrabi, R.; Blakemore, D.; Nafria, M.; Kestner, D.; Frampton, J.; Murphy, G.; et al. Fine-Tuning Mybl2 Is Required for Proper Mesenchymal-to-Epithelial Transition during Somatic Reprogramming. Cell Rep. 2018, 24, 1496–1511.e8. [Google Scholar] [CrossRef]
- Lemp, N.A.; Hiraoka, K.; Kasahara, N.; Logg, C.R. Cryptic transcripts from a ubiquitous plasmid origin of replication confound tests for cis-regulatory function. Nucleic Acids Res. 2012, 40, 7280–7290. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Herwig, A.; Osterhof, C.; Keppner, A.; Maric, D.; Koay, T.W.; Mbemba-Nsungi, A.; Hoogewijs, D. Ectopic MYBL2-Mediated Regulation of Androglobin Gene Expression. Cells 2024, 13, 826. https://doi.org/10.3390/cells13100826
Herwig A, Osterhof C, Keppner A, Maric D, Koay TW, Mbemba-Nsungi A, Hoogewijs D. Ectopic MYBL2-Mediated Regulation of Androglobin Gene Expression. Cells. 2024; 13(10):826. https://doi.org/10.3390/cells13100826
Chicago/Turabian StyleHerwig, Antonia, Carina Osterhof, Anna Keppner, Darko Maric, Teng Wei Koay, Ambre Mbemba-Nsungi, and David Hoogewijs. 2024. "Ectopic MYBL2-Mediated Regulation of Androglobin Gene Expression" Cells 13, no. 10: 826. https://doi.org/10.3390/cells13100826




