Soil Microbial Community Characteristics and Their Effect on Tea Quality under Different Fertilization Treatments in Two Tea Plantations
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Tea Sampling and Analysis
2.3. Soil Sampling and Analysis
2.4. DNA Extraction and Library Construction
2.5. Quality Control of Raw Sequencing Data
2.6. Operational Taxonomic Units (OTUs) Cluster and Species Annotation
2.7. Alpha Diversity Analysis
2.8. Beta Diversity Analysis
3. Results
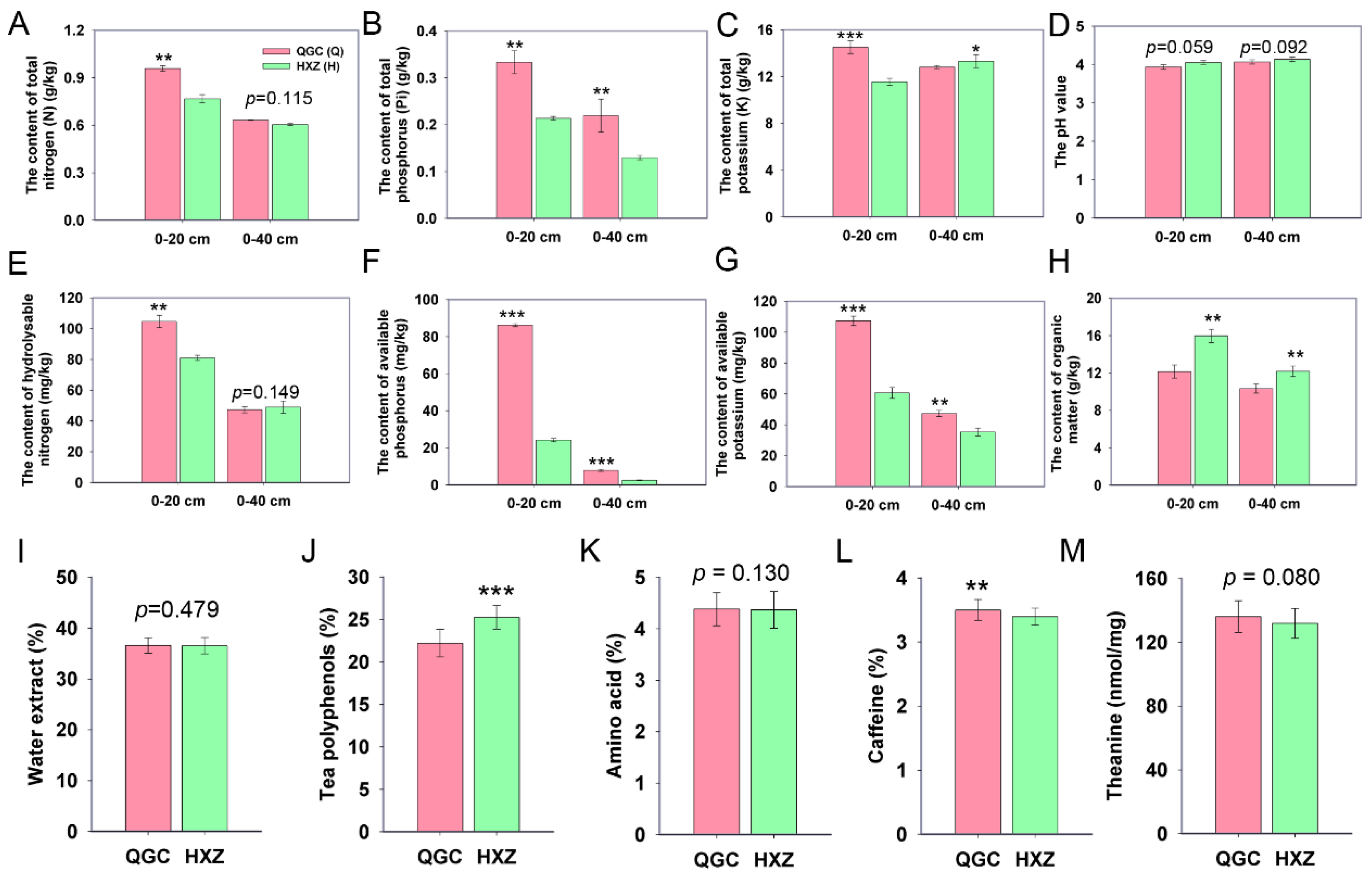
3.1. Chemical Characteristics of Soil and Tea Quality
3.2. Sequence Evaluation and Species Annotation
3.3. Alpha Diversity of the Soil Microbial Community
3.4. Beta Diversity of Microbial Composition
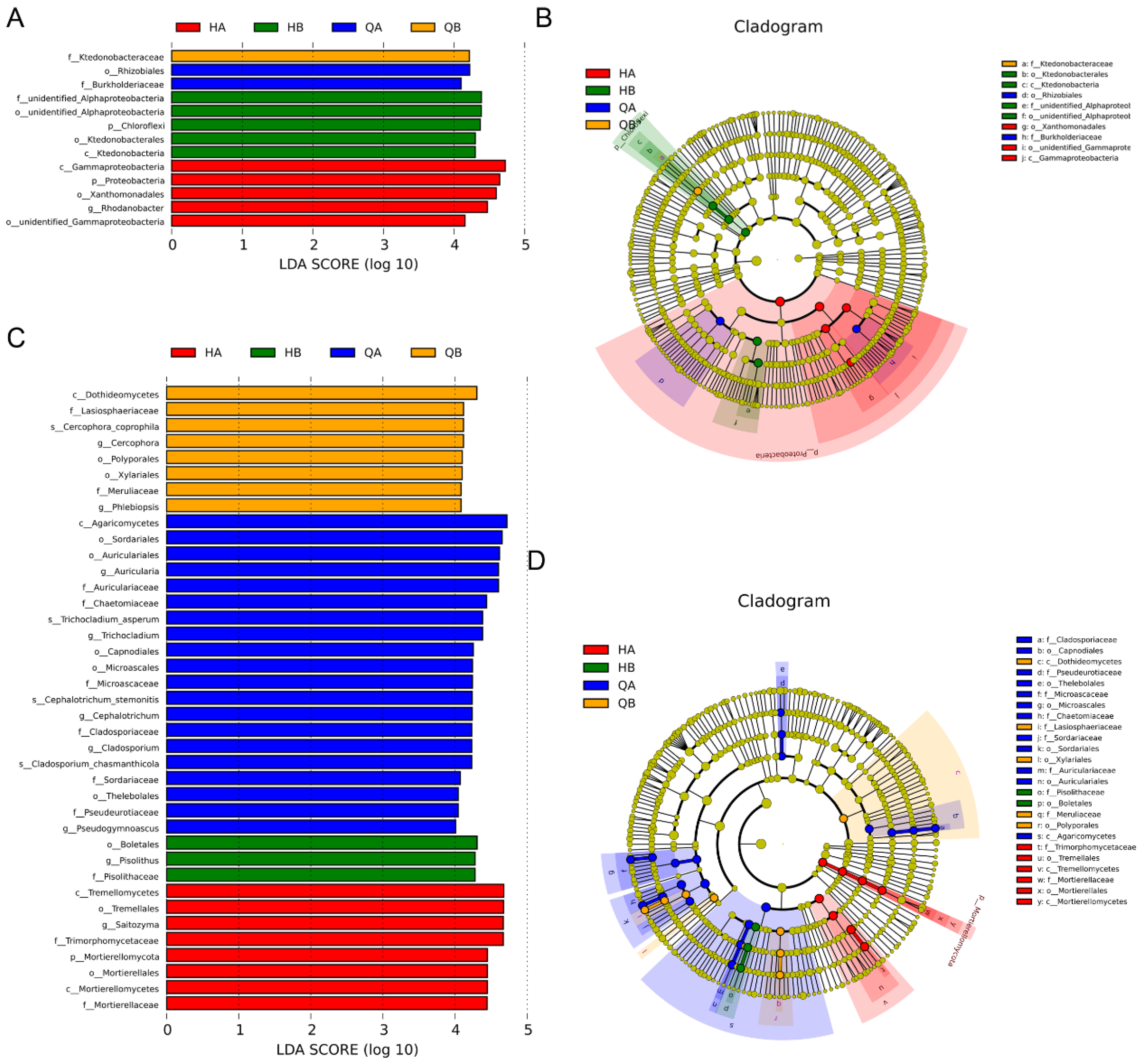
3.5. Bacterial Community Structure and Abundance
3.6. Fungal Community Structure and Abundance
3.7. Effect of Soil Properties on Microbial Communities
3.8. Network Analysis of Microbial Communities
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kumar, U.; Shahid, M.; Tripathi, R.; Mohanty, S.; Kumar, A.; Bhattacharyya, P.; Lal, B.; Gautam, P.; Raja, R.; Panda, B.B.; et al. Variation of functional diversity of soil microbial community in sub-humid tropical rice-rice cropping system under long-term organic and inorganic fertilization. Ecol. Indic. 2017, 73, 536–543. [Google Scholar] [CrossRef]
- Leifheit, E.F.; Veresoglou, S.D.; Lehmann, A.; Morris, E.K.; Rillig, M.C. Multiple factors influence the role of arbuscular mycorrhizal fungi in soil aggregation—A meta-analysis. Plant Soil 2013, 374, 523–537. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, H.; Dai, Y.; Tian, H.; Zhou, W.; Lv, J. Soil organic carbon transformation and dynamics of microorganisms under different organic amendments. Sci. Total Environ. 2021, 750, 141719. [Google Scholar] [CrossRef] [PubMed]
- Bergelson, J.; Mittelstrass, J.; Horton, M.W. Characterizing both bacteria and fungi improves understanding of the Arabidopsis root microbiome. Sci. Rep. 2019, 9, 24. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Geng, Y.; Liang, T. Optimization of reduced chemical fertilizer use in tea gardens based on the assessment of related environmental and economic benefits. Sci. Total Environ. 2020, 713, 136439. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, J. Spatiotemporal Changes of Chemical Fertilizer Application and Its Environmental Risks in China from 2000 to 2019. Int. J. Environ. Res. Public Health 2021, 18, 11911. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-R.; Luo, S.-Q.; Peng, Y.-J.; Jin, C.-Z.; Liu, D.-C. Effect of long-term application of bioorganic fertilizer on the soil property and bacteria in rice paddy. AMB Express 2023, 13, 60. [Google Scholar] [CrossRef]
- Wu, M.; Qin, H.; Chen, Z.; Wu, J.; Wei, W. Effect of long-term fertilization on bacterial composition in rice paddy soil. Biol. Fertil. Soils 2011, 47, 397–405. [Google Scholar] [CrossRef]
- Khan, K.S.; Ali, M.M.; Naveed, M.; Rehmani, M.I.A.; Shafique, M.W.; Ali, H.M.; Abdelsalam, N.R.; Ghareeb, R.Y.; Feng, G. Co-application of organic amendments and inorganic P increase maize growth and soil carbon, phosphorus availability in calcareous soil. Front. Environ. Sci. 2022, 10, 949371. [Google Scholar] [CrossRef]
- Tu, C.; Ristaino, J.B.; Hu, S. Soil microbial biomass and activity in organic tomato farming systems: Effects of organic inputs and straw mulching. Soil Biol. Biochem. 2006, 38, 247–255. [Google Scholar] [CrossRef]
- Hartmann, M.; Six, J. Soil structure and microbiome functions in agroecosystems. Nat. Rev. Earth Environ. 2022, 4, 4–18. [Google Scholar] [CrossRef]
- Zhu, L.; Yan, H.; Zhou, G.-S.; Jiang, C.-H.; Liu, P.; Yu, G.; Guo, S.; Wu, Q.-N.; Duan, J.-A. Insights into the mechanism of the effects of rhizosphere microorganisms on the quality of authentic Angelica sinensis under different soil microenvironments. BMC Plant Biol. 2021, 21, 285. [Google Scholar] [CrossRef] [PubMed]
- Finch, S.; Billiald, H.; Collier, R.H. Companion planting—Do aromatic plants disrupt host-plant finding by the cabbage root fly and the onion fly more effectively than non-aromatic plants? Entomol. Exp. Et. Appl. 2003, 109, 183–195. [Google Scholar] [CrossRef]
- Moore, K.J.; Anex, R.P.; Elobeid, A.E.; Fei, S.; Flora, C.B.; Goggi, A.S.; Jacobs, K.L.; Jha, P.; Kaleita, A.L.; Karlen, D.L.; et al. Regenerating Agricultural Landscapes with Perennial Groundcover for Intensive Crop Production. Agronomy 2019, 9, 458. [Google Scholar] [CrossRef]
- Chen, L.; Li, D.; Shao, Y.; Adni, J.; Wang, H.; Liu, Y.; Zhang, Y. Comparative Analysis of Soil Microbiome Profiles in the Companion Planting of White Clover and Orchard Grass Using 16S rRNA Gene Sequencing Data. Front. Plant Sci. 2020, 11, 538311. [Google Scholar] [CrossRef] [PubMed]
- Guertal, E.A.; Green, B.D. Evaluation of organic fertilizer sources for south-eastern (USA) turfgrass maintenance. Acta Agric. Scand. Sect. B—Soil. Plant Sci. 2012, 62 (Suppl. S1), 130–138. [Google Scholar] [CrossRef]
- Lazcano, C.; Gómez-Brandón, M.; Revilla, P.; Domínguez, J. Short-term effects of organic and inorganic fertilizers on soil microbial community structure and function. Biol. Fertil. Soils 2012, 49, 723–733. [Google Scholar] [CrossRef]
- Shi, Z.; Yang, Y.; Fan, Y.; He, Y.; Li, T. Dynamic Responses of Rhizosphere Microorganisms to Biogas Slurry Combined with Chemical Fertilizer Application during the Whole Life Cycle of Rice Growth. Microorganisms 2023, 11, 1755. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.; Hao, F.; He, W.; Ran, Q.; Nie, G.; Huang, L.; Wang, X.; Yuan, S.; Xu, W.; Zhang, X. Effect of Biogas Slurry on the Soil Properties and Microbial Composition in an Annual Ryegrass-Silage Maize Rotation System over a Five-Year Period. Microorganisms 2024, 12, 716. [Google Scholar] [CrossRef] [PubMed]
- Ling, N.; Zhu, C.; Xue, C.; Chen, H.; Duan, Y.; Peng, C.; Guo, S.; Shen, Q. Insight into how organic amendments can shape the soil microbiome in long-term field experiments as revealed by network analysis. Soil Biol. Biochem. 2016, 99, 137–149. [Google Scholar] [CrossRef]
- García-Gil, J.C.; Plaza, C.; Soler-Rovira, P.; Polo, A. Long-term effects of municipal solid waste compost application on soil enzyme activities and microbial biomass. Soil Biol. Biochem. 2000, 32, 1907–1913. [Google Scholar] [CrossRef]
- Luan, H.; Gao, W.; Huang, S.; Tang, J.; Li, M.; Zhang, H.; Chen, X.; Masiliunas, D. Substitution of manure for chemical fertilizer affects soil microbial community diversity, structure and function in greenhouse vegetable production systems. PLoS ONE 2020, 15, e0214041. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Li, J.; Yao, L. Soil microbial community and physicochemical properties together drive soil organic carbon inCunninghamia lanceolataplantations of different stand ages. PeerJ 2022, 10, e13873. [Google Scholar] [CrossRef]
- Zahid, M.S.; Hussain, M.; Song, Y.; Li, J.; Guo, D.; Li, X.; Song, S.; Wang, L.; Xu, W.; Wang, S. Root-Zone Restriction Regulates Soil Factors and Bacterial Community Assembly of Grapevine. Int. J. Mol. Sci. 2022, 23, 15628. [Google Scholar] [CrossRef]
- Zhao, T.; Li, C.; Wang, S.; Song, X. Green Tea (Camellia sinensis): A Review of Its Phytochemistry, Pharmacology, and Toxicology. Molecules 2022, 27, 3909. [Google Scholar] [CrossRef]
- Wang, W.; Min, Q.; Sardans, J.; Wang, C.; Asensio, D.; Bartrons, M.; Peñuelas, J. Organic Cultivation of Jasmine and Tea Increases Carbon Sequestration by Changing Plant and Soil Stoichiometry. Agron. J. 2016, 108, 1636–1648. [Google Scholar] [CrossRef]
- Supriyadi, S.; Vera, I.L.P.; Purwanto, P. Soil Quality at Rice Fields with Organic, Semi-organic and Inorganic Management in Wonogiri Regency, Indonesia. Caraka Tani J. Sustain. Agric. 2021, 36, 259–269. [Google Scholar] [CrossRef]
- Deka, N.; Goswami, K. Organic cultivation and sustainable value chain development for tea smallholders: Findings from Assam, India. Sustain. Prod. Consum. 2022, 32, 562–579. [Google Scholar] [CrossRef]
- Lin, W.; Lin, M.; Zhou, H.; Wu, H.; Li, Z.; Lin, W. The effects of chemical and organic fertilizer usage on rhizosphere soil in tea orchards. PLoS ONE 2019, 14, e0217018. [Google Scholar] [CrossRef]
- Fei, T.; Fei, J.; Huang, F.; Xie, T.; Xu, J.; Zhou, Y.; Yang, P. The anti-aging and anti-oxidation effects of tea water extract in Caenorhabditis elegans. Exp. Gerontol. 2017, 97, 89–96. [Google Scholar] [CrossRef]
- Zhu, B.; Chen, L.B.; Lu, M.; Zhang, J.; Han, J.; Deng, W.W.; Zhang, Z.Z. Caffeine Content and Related Gene Expression: Novel Insight into Caffeine Metabolism in Camellia Plants Containing Low, Normal, and High Caffeine Concentrations. J. Agric. Food Chem. 2019, 67, 3400–3411. [Google Scholar] [CrossRef]
- Rai, K.P.; Rijal, S.K.; Shrestha, S.; Pokhrel, P. A simple HPLC Method for the Determination of Caffeine Content in Tea and Coffee. J. Food Sci. Technol. Nepal. 2016, 9, 74–78. [Google Scholar] [CrossRef]
- Gallardo, J.F.; Saavedra, J.; Martin-Patino, T.; Millan, A. Soil organic matter determination. Commun. Soil Sci. Plant Anal. 2008, 18, 699–707. [Google Scholar] [CrossRef]
- Bremner, J.M. Determination of nitrogen in soil by the Kjeldahl method. J. Agric. Sci. 2009, 55, 11–33. [Google Scholar] [CrossRef]
- Chen, B.; Yang, H.; Song, W.; Liu, C.; Xu, J.; Zhao, W.; Zhou, Z. Effect of N fertilization rate on soil alkali-hydrolyzable N, subtending leaf N concentration, fiber yield, and quality of cotton. Crop. J. 2016, 4, 323–330. [Google Scholar] [CrossRef]
- De Silva, C.S.; Koralage, I.S.A.; Weerasinghe, P.; Silva, N.R.N. The Determination of Available Phosphorus in Soil: A Quick and Simple Method. OUSL J. 2015, 8, 1–17. [Google Scholar] [CrossRef]
- Beng, K.C.; Corlett, R.T. Amplicon sequencing dataset of soil fungi and associated environmental variables collected in karst and non-karst sites across Yunnan province, southwest China. Data Brief. 2019, 27, 104575. [Google Scholar] [CrossRef] [PubMed]
- Pecundo, M.H.; Chang, A.C.G.; Chen, T.; Dela Cruz, T.E.E.; Ren, H.; Li, N. Full-Length 16S rRNA and ITS Gene Sequencing Revealed Rich Microbial Flora in Roots of Cycas spp. in China. Evol. Bioinform. Online 2021, 17, 1176934321989713. [Google Scholar] [CrossRef] [PubMed]
- Kao, W.C.; Stevens, K.; Song, Y.S. BayesCall: A model-based base-calling algorithm for high-throughput short-read sequencing. Genome Res. 2009, 19, 1884–1895. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahe, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Willis, A.D. Rarefaction, Alpha Diversity, and Statistics. Front. Microbiol. 2019, 10, 2407. [Google Scholar] [CrossRef] [PubMed]
- Ceccarani, C.; Foschi, C.; Parolin, C.; D’Antuono, A.; Gaspari, V.; Consolandi, C.; Laghi, L.; Camboni, T.; Vitali, B.; Severgnini, M.; et al. Diversity of vaginal microbiome and metabolome during genital infections. Sci. Rep. 2019, 9, 14095. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Lladser, M.E.; Knights, D.; Stombaugh, J.; Knight, R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011, 5, 169–172. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Gong, W. The role of chemical and organic fertilizers on yield, yield variability and carbon sequestration—Results of a 19-year experiment. Plant Soil 2010, 331, 471–480. [Google Scholar] [CrossRef]
- Cheng, H.; Zhang, D.; Huang, B.; Song, Z.; Ren, L.; Hao, B.; Liu, J.; Zhu, J.; Fang, W.; Yan, D.; et al. Organic fertilizer improves soil fertility and restores the bacterial community after 1,3-dichloropropene fumigation. Sci. Total Environ. 2020, 738, 140345. [Google Scholar] [CrossRef]
- Li, Y.; Li, Z.; Arafat, Y.; Lin, W.; Jiang, Y.; Weng, B.; Lin, W. Characterizing rhizosphere microbial communities in long-term monoculture tea orchards by fatty acid profiles and substrate utilization. Eur. J. Soil Biol. 2017, 81, 48–54. [Google Scholar] [CrossRef]
- He, H.; Peng, M.; Hou, Z.; Li, J. Unlike chemical fertilizer reduction, organic fertilizer substitution increases soil organic carbon stock and soil fertility in wheat fields. J. Sci. Food Agric. 2023, 104, 2798–2808. [Google Scholar] [CrossRef]
- Zama, N.; Kirkman, K.; Mkhize, N.; Tedder, M.; Magadlela, A. Soil Acidification in Nutrient-Enriched Soils Reduces the Growth, Nutrient Concentrations, and Nitrogen-Use Efficiencies of Vachellia sieberiana (DC.) Kyal. & Boatwr Saplings. Plants 2022, 11, 3564. [Google Scholar] [CrossRef]
- Nakamaru, Y.M.; Matsuda, R.; Sonoda, T. Environmental risks of organic fertilizer with increased heavy metals (Cu and Zn) to aquatic ecosystems adjacent to farmland in the northern biosphere of Japan. Sci. Total Environ. 2023, 884, 163861. [Google Scholar] [CrossRef]
- Chen, L.; Li, X.; Peng, Y.; Xiang, P.; Zhou, Y.; Yao, B.; Zhou, Y.; Sun, C. Co-application of biochar and organic fertilizer promotes the yield and quality of red pitaya (Hylocereus polyrhizus) by improving soil properties. Chemosphere 2022, 294, 133619. [Google Scholar] [CrossRef]
- Jee, H.J.; Lee, S.G.; Bormate, K.J.; Jung, Y.S. Effect of Caffeine Consumption on the Risk for Neurological and Psychiatric Disorders: Sex Differences in Human. Nutrients 2020, 12, 3080. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Cui, S.; Ma, J.; Wu, D.; Ye, Z.; Liu, D. Effects of Shellfish and Organic Fertilizer Amendments on Soil Nutrients and Tea Yield and Quality. Toxics 2023, 11, 262. [Google Scholar] [CrossRef]
- Ye, J.; Wang, Y.; Kang, J.; Chen, Y.; Hong, L.; Li, M.; Jia, Y.; Wang, Y.; Jia, X.; Wu, Z.; et al. Effects of Long-Term Use of Organic Fertilizer with Different Dosages on Soil Improvement, Nitrogen Transformation, Tea Yield and Quality in Acidified Tea Plantations. Plants 2022, 12, 122. [Google Scholar] [CrossRef] [PubMed]
- Manzoor; Ma, L.; Ni, K.; Ruan, J. Influence of Organic and Inorganic Fertilizers on Tea Growth and Quality and Soil Properties of Tea Orchards’ Top Rhizosphere Soil. Plants 2024, 13, 207. [Google Scholar] [CrossRef]
- Yan, Y.; Kuramae, E.E.; de Hollander, M.; Klinkhamer, P.G.; van Veen, J.A. Functional traits dominate the diversity-related selection of bacterial communities in the rhizosphere. ISME J. 2017, 11, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Guan, D.; Ma, M.; Jiang, X.; Fan, F.; Meng, F.; Li, L.; Zhao, B.; Zhao, Y.; Cao, F.; et al. Long-term fertilization coupled with rhizobium inoculation promotes soybean yield and alters soil bacterial community composition. Front. Microbiol. 2023, 14, 1161983. [Google Scholar] [CrossRef]
- Lauber, C.L.; Strickland, M.S.; Bradford, M.A.; Fierer, N. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. Biochem. 2008, 40, 2407–2415. [Google Scholar] [CrossRef]
- Li, Y.C.; Li, Z.; Li, Z.W.; Jiang, Y.H.; Weng, B.Q.; Lin, W.X. Variations of rhizosphere bacterial communities in tea (Camellia sinensis L.) continuous cropping soil by high-throughput pyrosequencing approach. J. Appl. Microbiol. 2016, 121, 787–799. [Google Scholar] [CrossRef] [PubMed]
- de Sousa Lopes, L.; Mendes, L.W.; Antunes, J.E.L.; de Souza Oliveira, L.M.; Melo, V.M.M.; de Araujo Pereira, A.P.; da Costa, A.F.; de Paula Oliveira, J.; Martínez, C.R.; Figueiredo, M.d.V.B.; et al. Distinct bacterial community structure and composition along different cowpea producing ecoregions in Northeastern Brazil. Sci. Rep. 2021, 11, 831. [Google Scholar] [CrossRef]
- Mendes, R.; Kruijt, M.; de Bruijn, I.; Dekkers, E.; van der Voort, M.; Schneider, J.H.; Piceno, Y.M.; DeSantis, T.Z.; Andersen, G.L.; Bakker, P.A.; et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Wang, Y.; Jin, X.; Zhao, Y.; Yan, H.; Zhang, H.; Zhou, X.; Lu, G.; Deng, Y. Application of Organic Fertilizer Changes the Rhizosphere Microbial Communities of a Gramineous Grass on Qinghai–Tibet Plateau. Microorganisms 2022, 10, 1148. [Google Scholar] [CrossRef] [PubMed]
- Jankowiak, J.G.; Gobler, C.J. The Composition and Function of Microbiomes Within Microcystis Colonies Are Significantly Different Than Native Bacterial Assemblages in Two North American Lakes. Front. Microbiol. 2020, 11, 1016. [Google Scholar] [CrossRef] [PubMed]
- Spain, A.M.; Krumholz, L.R.; Elshahed, M.S. Abundance, composition, diversity and novelty of soil Proteobacteria. ISME J. 2009, 3, 992–1000. [Google Scholar] [CrossRef]
- Li, Q.; Bao, Z.; Tang, K.; Feng, H.; Tu, W.; Li, L.; Han, Y.; Cao, M.; Zhao, C. First two mitochondrial genomes for the order Filobasidiales reveal novel gene rearrangements and intron dynamics of Tremellomycetes. IMA Fungus 2022, 13, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Gao, Y.; Jia, X.; Wang, M.; Ding, J.; Cheng, L.; Bao, F.; Wu, B. Network analysis reveals the strengthening of microbial interaction in biological soil crust development in the Mu Us Sandy Land, northwestern China. Soil Biol. Biochem. 2020, 144, 107782. [Google Scholar] [CrossRef]
- Gundlapally, S.R.; Garcia-Pichel, F. The community and phylogenetic diversity of biological soil crusts in the Colorado Plateau studied by molecular fingerprinting and intensive cultivation. Microb. Ecol. 2006, 52, 345–357. [Google Scholar] [CrossRef]
- Semenov, M.V.; Krasnov, G.S.; Semenov, V.M.; van Bruggen, A. Mineral and Organic Fertilizers Distinctly Affect Fungal Communities in the Crop Rhizosphere. J. Fungi 2022, 8, 251. [Google Scholar] [CrossRef]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lei, Y.; Ding, D.; Duan, J.; Luo, Y.; Huang, F.; Kang, Y.; Chen, Y.; Li, S. Soil Microbial Community Characteristics and Their Effect on Tea Quality under Different Fertilization Treatments in Two Tea Plantations. Genes 2024, 15, 610. https://doi.org/10.3390/genes15050610
Lei Y, Ding D, Duan J, Luo Y, Huang F, Kang Y, Chen Y, Li S. Soil Microbial Community Characteristics and Their Effect on Tea Quality under Different Fertilization Treatments in Two Tea Plantations. Genes. 2024; 15(5):610. https://doi.org/10.3390/genes15050610
Chicago/Turabian StyleLei, Yu, Ding Ding, Jihua Duan, Yi Luo, Feiyi Huang, Yankai Kang, Yingyu Chen, and Saijun Li. 2024. "Soil Microbial Community Characteristics and Their Effect on Tea Quality under Different Fertilization Treatments in Two Tea Plantations" Genes 15, no. 5: 610. https://doi.org/10.3390/genes15050610




