Cloning and Function Analysis of the CsTAU1 in Response to Salt–Alkali Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Cloning and Sequence Analysis of CsTAU1
2.3. Subcellular Localization of CsTAU1
2.4. Relative Expression of CsTAU1 in Different Tissues of Cucumber
2.5. CsTAU1 Expression under Different External Factors
2.6. Genetic Transformation of Cucumber by CsTAU1
2.7. Measurement of Physiological and Biochemical Indicators
2.8. Statistical Analysis
3. Results
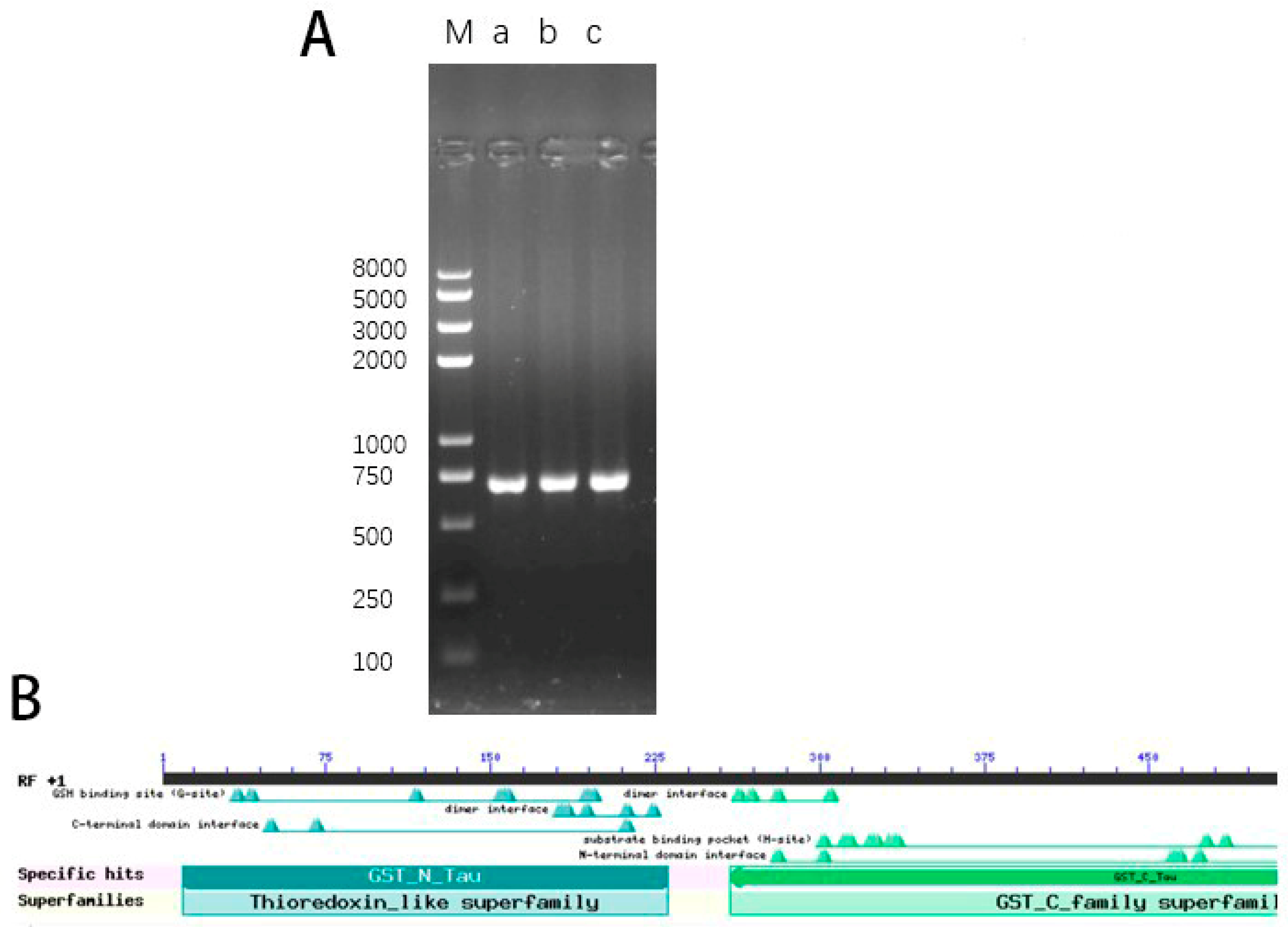
3.1. Cloning and Bioinformatics Analysis of CsTAU1
3.2. Subcellular Localization of the CsTAU1
3.3. Phylogenetic Tree of CsTAU1
3.4. Relative Expression of CsTAU1 in Different Tissues of Cucumber under Salt–Alkali Stress
3.5. Analysis of CsTAU1 Expression under Different External Factors
3.6. Obtaining CsTAU1 Overexpressed Resistant Plants
3.7. Performance of CsTAU1 Overexpressing Plants under Salt–Alkali Stress
3.8. CsTAU1 Reduces the Metabolic Pathway of Salt–Alkali Stress
3.9. Expression of Key Genes in the Glutathione Pathway in Overexpressing Plants
3.10. The Effect of Salt–Alkali Stress on Physiological Indicators of CsTAU1-Overexpressing Cucumbers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, S.R.; Huang, Y.X. Research progress on the improvement and management of saline alkali land in the Songnen Plain. Soil Crops 2023, 12, 206–217. [Google Scholar]
- Sun, X.J.; Jiang, Y.Q.; Hu, Z.; Li, H.B.; Pang, B.S.; Zhang, F.T.; Zhang, S.Q.; Zhang, H. Identification and evaluation of salt tolerance in wheat germplasm resources during seedling stage. Acta Agron. Sin. 2023, 49, 1132–1139. [Google Scholar]
- Bai, J.Z.; Zhao, Y.; Wu, F.Z. The effect of salt alkali stress on the structure and abundance of bacterial and fungal communities in the rhizosphere soil of cucumber grafted seedlings. Chin. J. Eco-Agric. 2017, 25, 1626–1635. [Google Scholar]
- Han, Z.D.; Sun, H.F.; Feng, L.Y.; Zhang, L.; Jia, H.C.; Yan, X.F.; Lu, W.C. Preliminary study on pH regulation of soybean salt alkali tolerance identification pool. Heilongjiang Agric. Sci. 2023, 6, 1–5. [Google Scholar]
- Liu, F.J.; Zhang, X.W.; Cai, B.B.; Pan, D.Y.; Fu, X.; Bi, H.G.; Ai, X.Z. Physiological response and transcription profiling analysis reveal the role of glutathione in H2S-induced chilling stress tolerance of cucumber seedlings. Plant Sci. 2020, 219, 110363. [Google Scholar] [CrossRef] [PubMed]
- Kuźniak, E.; Głowacki, R.; Chwatko, G.; Kopczewski, T.; Wielanek, M.; Gajewska, E. Involvement of ascorbate, glutathione, protein S-thiolation and salicylic acid in benzothiadiazole-inducible defence response of cucumber against Pseudomonas syringae pv lachrymans. Physiol. Mol. Plant Pathol. 2014, 86, 89–97. [Google Scholar] [CrossRef]
- Hong, J.Y.; Son, H.S.; Hong, S.P.; Yi, S.H.; Kang, S.H.; Lee, M.K.; Paik, H.D. Production of β-glucan, glutathione, and glutathione derivatives by probiotic Saccharomyces cerevisiae isolated from cucumber jangajji. LWT 2018, 100, 114–118. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Hossain, M.A.; Silva, D.; Fujita, M. Plant response and tolerance to abiotic oxidative stress: Antioxidant defense is a key factor. In Cropstress and Its Management: Perspectives and Strategies; Venkateswarlu, B., Shanker, A.K., Shanker, C., Maheswari, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 261–315. [Google Scholar]
- Gao, H.S. Functional Study of OsCYP714D1 and PtGSTF1 in improving poplar growth and salt resistance. Master’s Thesis, Ludong University, Yantai, China, March 2024. [Google Scholar]
- Wang, Y.; Jia, B.C.; Wu, X.L.; Zhang, D.L.; Zhu, J.; Tian, X.Q.; Chen, S.H.; Guo, S.L. Identification and expression pattern analysis of the CqNHX gene family in quinoa. J. Yantai Univ. 2020, 33, 270–275. [Google Scholar]
- Liang, G.W.; Li, Z.Q.; Zhou, B.J.; Chang, M.M.; Chen, T.; Chen, P. Cloning and expression analysis of glutathione reductase gene (HcGR) from kenaf under salt stress. J. South. Agric. 2020, 51, 2412–2419. [Google Scholar]
- Cimini, S.; Locato, V.; Giacinti, V.; Molinari, M.; De, G. A multifactorial regulation of glutathione metabolism behind salt tolerance in rice. Antioxidants 2022, 11, 1114. [Google Scholar] [CrossRef]
- Zhao, X.L.; Xu, G.F.; Zhao, C.F.; Sun, S.A.; Zhao, X.Z. The effect of garlic on the activities of glutathione S-transferase, glutathione peroxidase, and glutathione content in mice. Mod. Prev. Med. 1998, 10, 3. [Google Scholar]
- Altaf, M.A.; Shahid, R.; Ren, M.X.; Altaf, M.M.; Khan, L.U.; Shahid, S. Melatonin alleviates salt damage in tomato seedling: A root architecture system, photosynthetic capacity, ion homeostasis, and antioxidant enzymes analysis. Sci. Hortic. 2021, 285, 110145. [Google Scholar] [CrossRef]
- Zhang, F.; Zhang, J.M.; Li, Q.F.; Yang, Y.; Sheng, Y.Y. Exploring Candidate Genes and Regulatory Mechanisms for Salt–Alkali Tolerance in Cucumber. Agronomy 2024, 14, 543. [Google Scholar] [CrossRef]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient geneexpression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.Q.; Gao, Y.M.; Du, Y.L.; Zhang, Y.X.; Du, J.D.; Zhang, W.H.; Pan, S.Y. Identification of salt and alkali resistant soybean germplasm resources. Chin. J. Oil Crop Sci. 2021, 43, 1016–1024. [Google Scholar]
- Wan, H.; Zhao, Z.; Qian, C.; Sui, Y.; Malik, A.A.; Chen, J. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal. Biochem. 2010, 399, 257–261. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Li, C.X.; Zhang, M.L.; Qi, N.N.; Liu, H.W.; Zhao, Z.X.; Huang, P.P.; Liao, W.B. Abscisic acid induces adventitious rooting in cucumber (Cucumis sativus L.) by enhancing sugar synthesis. Plants 2022, 11, 2354. [Google Scholar] [CrossRef]
- Chen, S.; Songkumarn, P.; Liu, J.; Wang, G.L. A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol. 2009, 150, 1111–1121. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Z.; Zhang, X.L. A GAMYB homologue CsGAMYB1 regulates sex expression of cucumber via an ethylene-independent pathway. J. Exp. Bot. 2014, 65, 3201–3213. [Google Scholar] [CrossRef]
- Zhao, S.J.; Cang, J. Guidance for Plant Physiology Experiments; China Agriculture Press: Beijing, China, 2016. [Google Scholar]
- Lea, U.S.; Slimestad, R.; Smedvig, P. Nitrogen deficiency enhances expression of specific MYB and bHLH transcription factors and accumulation of end products in the flavonoid pathway. Planta 2007, 225, 1245–1253. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Wang, J.F.; Hou, W.P.; Malik, K.; Zhao, C.Z.; Niu, X.L.; Liu, Y.L.; Huang, R.; Li, C.J.; Nan, Z.B. Elucidating the molecular mechanisms by which seed-borne endophytic fungi, epichloë gansuensis, increases the tolerance of achnatherum inebrians to NaCl stress. Int. J. Mol. Sci. 2021, 22, 13191. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.J.; Mu, M.; Lu, X.K.; Yin, Z.J.; Wang, D.L.; Wang, J.J.; Fan, W.L.; Wang, S.; Ye, W.W. Expression and functional analysis of salt tolerance related gene GhVP in cotton. Cotton Sci. 2016, 28, 122–128. [Google Scholar]
- Peng, Y.Q.; Cao, H.S.; Cui, L.J.; Wang, Y.; Wei, L.X.; Geng, S.Y.; Yang, L.; Huang, Y.; Bie, Z.L. CmoNAC1 in pumpkin rootstocks improves salt tolerance of grafted cucumbers by binding to the promoters of CmoRBOHD1, CmoNCED6, CmoAKT1;2 and CmoHKT1;1 to regulate H2O2, ABA signaling and K+/Na+ homeostasis. Hortic. Res. 2023, 10, 157. [Google Scholar] [CrossRef]
- Hao, J.H.; Yi, Y.; Shang, Q.M.; Dong, C.J.; Zhang, Z. Effect of exogenous salicylic acid on nitrogen assimilation of cucumber seedling under drought stress. Acta Hortic. Sin. 2012, 39, 81–90. [Google Scholar]
- Daniel, M.M.; Said, E.; Pa, R.N. Catalysis within the lipid bilayer-structure and mechanism of the MAPEG family of integral membrane proteins. Curr. Opin. Struct. Biol. 2008, 18, 442–449. [Google Scholar]
- Dröge, W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002, 82, 47–95. [Google Scholar] [CrossRef] [PubMed]
- Carlo, W.T.; Van, R.; Martin, G.; Schroers, J. The peroxisomal NAD carrier from arabidopsis imports NAD in exchange with AMP. Plant Physiol. 2016, 171, 2127–2139. [Google Scholar]
- Fulda, M.; Schnurr, J.; Abbadi, A.; Heinz, E.; Browse, J. Peroxisomal Acyl-CoA synthetase activity is essential for seedling development in Arabidopsis thaliana. Plant Cell 2004, 16, 394–405. [Google Scholar] [CrossRef]
- Zhang, H.B.; Mu, D.; Li, Y.S.; Li, X.L.; Yan, X. Glutathione S-transferase activity facilitates rice tolerance to the barnyard grass root exudate DIMBOA. BMC Plant Biol. 2024, 24, 117. [Google Scholar] [CrossRef]
- Wang, X.; Chen, K.; Zhou, M.; Gao, Y.; Huang, H.; Liu, C.; Fan, Y.; Fan, Z.; Wang, Y.; Li, X. GmNAC181 promotes symbiotic nodulation and salt tolerance of nodulation by directly regulating GmNINa expression in soybean. New Phytol. 2022, 236, 656–670. [Google Scholar] [CrossRef] [PubMed]
- Farkas, M.H.; Berry, J.O. Chlortetracycline detoxification in maize via induction of Glutathione S-transferases after antibiotic exposure. Environ. Sci Technol. 2007, 41, 1450–1456. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.C.; Fu, Y.R.; Guo, Y.Q.; Ma, Y.W.; Li, S.X.; Lin, J.X.; Wang, J.H. Comparative effects of water potential stress induced by salt, alkali and drought on photosynthetic electron transport and apparatus in Hordeum jubatum seedlings. Crop Pasture Sci. 2022, 74, 324–333. [Google Scholar] [CrossRef]
- Álvarez-Robles, M.J.; Bernal, M.P.; Sánchez-Guerrero, A.; Sevilla, F.; Clemente, R. Major As species, lipid peroxidation and protein carbonylation in rice plants exposed to increasing as(V) concentrations. Heliyon 2020, 8, 04703. [Google Scholar] [CrossRef] [PubMed]
- Gémes, K.; Poór, P.; Horváth, E.; Kolbert, Z.; Szopkó’s, D.; Szepesi, A. Cross-talk between salicylic acid and NaCl-generated reactive oxygen species and nitric oxide in tomato during acclimation to high salinity. Physiol. Plant. 2011, 142, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Jain, S.K.; Tomar, B.S.; Anand, A.; Singh, J.; Sagar, V.; Kumar, R.; Singh, V.; Chaubey, T.; Abd-Elsalam, K.A.; et al. Impact of Foliar Application of ZnO2 and Fe3O4 Nanoparticles on Seed Yield and Physio-Biochemical Parameters of Cucumber (Cucumis sativus L.) Seed under Open Field and Protected Environment vis a vis during Seed Germination. Plants 2022, 11, 3211. [Google Scholar] [CrossRef]
- Priyanka, P.; Kuldeep, Y.; Krishan, K.; Narender, S. Effect of Gibberellic Acid and Potassium Foliar Sprays on Productivity and Physiological and Biochemical Parameters of Parthenocarpic Cucumber cv. ‘Seven Star F1’. J. Hortic. Res. 2016, 24, 93–100. [Google Scholar]










Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, F.; Li, D.; Sa, R.; Wang, L.; Sheng, Y. Cloning and Function Analysis of the CsTAU1 in Response to Salt–Alkali Stress. Genes 2024, 15, 613. https://doi.org/10.3390/genes15050613
Zhang F, Li D, Sa R, Wang L, Sheng Y. Cloning and Function Analysis of the CsTAU1 in Response to Salt–Alkali Stress. Genes. 2024; 15(5):613. https://doi.org/10.3390/genes15050613
Chicago/Turabian StyleZhang, Fan, Dandan Li, Rina Sa, Ling Wang, and Yunyan Sheng. 2024. "Cloning and Function Analysis of the CsTAU1 in Response to Salt–Alkali Stress" Genes 15, no. 5: 613. https://doi.org/10.3390/genes15050613




