Factor Analysis of Genetic Parameters for Body Conformation Traits in Dual-Purpose Simmental Cattle
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Phenotypic and Pedigree Data
2.2. Genotype Data
2.3. Genetic Connectedness
2.4. Variance Component Estimates for Body Conformation Traits
2.5. Factor Analysis
2.6. Estimation of Genetic Parameters Using Factor Analysis
3. Results
3.1. Phenotype
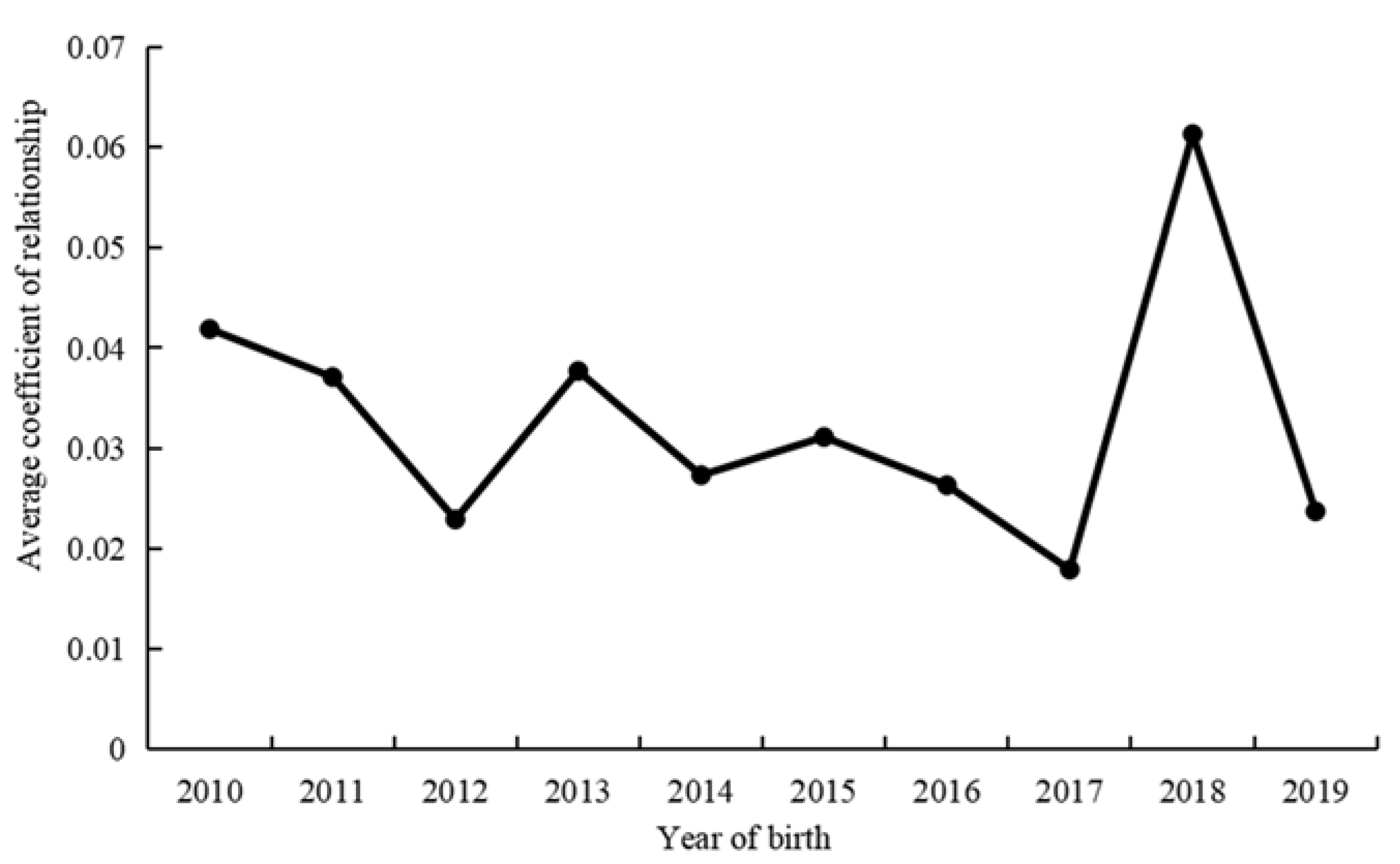
3.2. Genetic Connectedness
3.3. Heritability of Conformation Traits
3.4. Factor Analysis
3.5. Heritability of Factor Scores
3.6. Correlations between EBV of Composite Traits and EBV of Factor Scores
4. Discussion
4.1. Phenotype
4.2. Heritability
4.3. Factor Analysis
4.4. Correlations between EBV of Composite Traits and EBV of Factor Scores
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Perišić, P.; Skalicki, Z.; Petrovic, M.; Bogdanović, V.; Ružić-Muslić, D. Simmental cattle breed in different production systems. Biotechnol. Anim. Husban 2009, 25, 315–326. [Google Scholar] [CrossRef]
- Amaya, A.; Garrick, D.; Martínez, R.; Cerón-Muñoz, M. Economic values for index improvement of dual-purpose Simmental cattle. Livest. Sci. 2020, 240, 104224. [Google Scholar] [CrossRef]
- Wei, C.; Luo, H.; Wang, Y.; Huang, X.; Zhang, M.; Zhang, X.; Wang, D.; Ge, J.; Xu, L.; Jiang, H.; et al. Analyses of the genetic relationships between lactose, somatic cell score, and growth traits in Simmental cattle. Animal 2021, 15, 100027. [Google Scholar] [CrossRef] [PubMed]
- Edel, C.; Schwarzenbacher, H.; Hamann, H.; Neuner, S.; Emmerling, R.; Götz, K. The German-Austrian genomic evaluation system for Fleckvieh (Simmental) cattle. Interbull Bull. 2011, 44, 152–156. [Google Scholar]
- Chen, W. Estimation of Breeding Values in Simmental Cattle Using Animal BLUP Model; Xinjiang Agricultural University: Urumqi, China, 2017. [Google Scholar]
- Zhu, B.; Li, J.; Wang, C.; Xu, L.; Chen, Y.; Gao, X.; Zhang, L.; Gao, H.; Li, J. Genetic parameter and genetic gain estimation for growth and development traits in Chinese Simmental beef cattle. Acta Vet. Zootech. Sin. 2020, 51, 1833–1844. [Google Scholar]
- Stefani, G.; El Faro, L.; Santana, M.; Tonhati, H. Association of longevity with type traits, milk yield and udder health in Holstein cows. Livest. Sci. 2018, 218, 1–7. [Google Scholar] [CrossRef]
- Zink, V.; Štípková, M.; Lassen, J. Genetic parameters for female fertility, locomotion, body condition score, and linear type traits in Czech Holstein cattle. J. Dairy Sci. 2011, 94, 5176–5182. [Google Scholar] [CrossRef]
- Gibson, K.D.; Dechow, C.D. Genetic parameters for yield, fitness, and type traits in US Brown Swiss dairy cattle. J. Dairy Sci. 2017, 101, 1251–1257. [Google Scholar] [CrossRef]
- Pérez-Cabal, M.; Alenda, R. Genetic relationships between lifetime profit and type traits in Spanish Holstein cows. J. Dairy Sci. 2003, 85, 3480–3491. [Google Scholar] [CrossRef]
- Török, E.; Komlosi, I.; Szőnyi, V.; Béla, B.; Mészáros, G.; Posta, J. Combinations of linear type tra, its affecting the longevity in Hungarian Holstein-Friesian cows. Animals 2021, 11, 3065. [Google Scholar] [CrossRef]
- Simčič, M.; Luštrek, B.; Štepec, M.; Logar, B.; Potočnik, K. Estimation of genetic parameters of type traits in first parity cows of the Autochthonous Cika cattle in Slovenia. Front. Genet. 2021, 12, 724058. [Google Scholar] [CrossRef] [PubMed]
- Vanraden, P.M.; Jensen, E.L.; Lawlor, T.; Funk, D.A. Prediction of transmitting abilities for Holstein type traits. J. Dairy. Sci. 1990, 73, 191–197. [Google Scholar] [CrossRef]
- Mazza, S.; Guzzo, N.; Sartori, C.; Berry, D.P.; Mantovani, R. Genetic parameters for linear type traits in the Rendena dual-purpose breed. J. Anim. Breed. Genet. 2013, 131, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Macciotta, N.; Cecchinato, A.; Mele, M.; Bittante, G. Use of multivariate factor analysis to define new indicator variables for milk composition and coagulation properties in Brown Swiss cows. J. Dairy Sci. 2012, 95, 7346–7354. [Google Scholar] [CrossRef] [PubMed]
- Corrales Alvarez, J.; Cerón-muñoz, M.; Cañas-Álvarez, J.; Herrera, R.; Calvo Cardona, S. Relationship between type traits and milk production in Holstein cows from Antioquia, Colombia. Rev. Mvz. Cordoba 2011, 16, 2507–2513. [Google Scholar]
- Vukasinovic, N.; Moll, J.; Künzi, N. Factor analysis for evaluating relationships between herd life and type traits in Swiss Brown cattle. Lives. Prod. Sci. 1997, 49, 227–234. [Google Scholar] [CrossRef]
- Mazza, S.; Guzzo, N.; Sartori, C.; Mantovani, R. Factor analysis for genetic evaluation of linear type traits in dual-purpose autochthonous breeds. Animal 2015, 10, 372–380. [Google Scholar] [CrossRef]
- Olasege, B.; Zhang, S.; Zhao, Q.; Liu, D.; Sun, H.; Wang, Q.; Ma, P.P.; Pan, Y. Genetic parameter estimates for body conformation traits using composite index, principal component, and factor analysis. J. Dairy Sci. 2019, 102, 5219–5229. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.; Bender, D.; Maller, J.; Sklar, P.; Bakker, P.; Daly, M.; et al. Plink: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Sargolzaei, M.; Iwaisaki, H.; Colleau, J.J. CFC: A tool for monitoring genetic diversity. In Proceedings of the 8th World Congress on Genetics Applied to Livestock Production, Belo Horizonte, Brazil, 13–18 August 2006; pp. 27–28. [Google Scholar]
- Misztal, I.; Tsuruta, S.; Lourenco, D.; Masuda, Y.; Aguilar, I.; Legarra, A.; Vitezica, Z. Manual for BLUPF90 Family of Programs; University of Georgia: Athens, Greece, 2016. [Google Scholar]
- Henderson, C. Best linear unbiased estimation and prediction under a selection model. Biometrics 1975, 31, 423–447. [Google Scholar] [CrossRef]
- Lourenco, D.; Legarra, A.; Tsuruta, S.; Masuda, Y.; Aguilar, I.; Misztal, I. Single-step genomic evaluations from theory to practice: Using SNP chips and sequence data in BLUPF90. Genes 2020, 11, 790. [Google Scholar] [CrossRef]
- Putz, A.; Tiezzi, F.; Maltecca, C.; Gray, K.; Kanuer, M. Variance component estimates for alternative litter size traits in swine. J. Anim Sci. 2015, 93, 5153–5163. [Google Scholar] [CrossRef] [PubMed]
- Russell, D. In search of underlying dimensions: The use (and abuse) of factor analysis in personality and social psychology bulletin. Pers. Soc. Psychol. B 2002, 28, 1629–1646. [Google Scholar] [CrossRef]
- Hair, J.F.J.; Black, W.C.; Babin, B.J.; Anderson, R.E. Exploratory Factor Analysis. In Multivariate Data Analysis: Pearson New International Edition, 7th ed.; Prentice Hall: Upper Saddle River, NJ, USA, 2014; p. 118. [Google Scholar]
- Strapáková, E.; Strapák, P.; Candrak, J.; Pavlík, I.; Dočkalová, K. Fleckscore system of exterior evaluation as a more accurate indirect predictor of longevity in Slovak Simmental dairy cows. Czech. J. Anim. Sci. 2021, 66, 487–494. [Google Scholar] [CrossRef]
- Zavadilová, L.; Němcová, E.; Štípková, M.; Bouška, J. Relationships between longevity and conformation traits in Czech Fleckvieh cows. Czech. J. Anim. Sci. 2009, 54, 387–394. [Google Scholar] [CrossRef]
- Mazza, S.; Sartori, C.; Mantovani, R. Genetic parameters of type traits in two strains of dual purpose autochthonous Valdostana cattle. Livest. Sci. 2015, 178, 35–42. [Google Scholar] [CrossRef]
- Tarres, J.; Liu, Z.; Reinhardt, F.; Reents, R.; Ducrocq, V. Binational evaluation of type traits from Germany and France with a single-trait MACE animal model. Animal 2009, 3, 925–932. [Google Scholar] [CrossRef]
- Novotný, L.; Frelich, J.; Beran, J.; Zavadilová, L. Genetic relationship between type traits, number of lactations initiated, and lifetime milk performance in Czech Fleckvieh cattle. Czech. J. Anim. Sci. 2017, 62, 2017–2501. [Google Scholar] [CrossRef]
- Pfeiffer, C.; Fuerst-Waltl, B.; Ducrocq, V.; Fuerst, C. Approximate multivariate genetic evaluation of functional longevity and type traits in Austrian Fleckvieh cattle. In Proceedings of the 10th World Congress on Genetics Applied to Livestock Production, Vancouver, BC, Canada, 17–22 August 2014; p. 386. [Google Scholar]
- Kern, E.; Cobuci, J.; Costa, C.; Mcmanus, C.; Braccini, N.J. Genetic association between longevity and linear type traits of Holstein cows. Sci. Agric. 2015, 72, 203–209. [Google Scholar] [CrossRef]
- Špehar, M.; Štepec, M.; Potočnik, K. Variance components estimation for type traits in Slovenian brown Swiss cattle. Acta Agric. Slov. 2012, 100, 107–115. [Google Scholar]
- Roveglia, C.; Niero, G.; Bobbo, T.; Penasa, M.; Finocchiaro, R.; Visentin, G.; Lopez-Villalobos, N.; Cassandro, M. Genetic parameters for linear type traits including locomotion in Italian Jersey cattle breed. Livest. Sci. 2019, 229, 131–136. [Google Scholar] [CrossRef]
- Campos, R.; Cobuci, J.; Costa, C.; Braccini, N.J. Genetic parameters for type traits in Holstein cows in Brazil. Rev. Bras. Zootecn. 2012, 41, 2150–2161. [Google Scholar] [CrossRef]
- Veerkamp, R.; Mulder, H.; Thompson, R.; Calus, M. Genomic and pedigree-based genetic parameters for scarcely recorded traits when some animals are genotyped. J. Dairy Sci. 2011, 94, 4189–4197. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Luo, H.; Zhao, B.; Tian, K.; Huang, X.; Wang, Y.; Fu, X.; Tian, Y.; Di, J.; Xu, X.; et al. The effect of integrating genomic information into genetic evaluations of Chinese Merino sheep. Animals 2020, 10, 569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naserkheil, M.; Lee, D.; Mehrban, H. Improving the accuracy of genomic evaluation for linear body measurement traits using single-step genomic best linear unbiased prediction in Hanwoo beef cattle. BMC Genet. 2020, 21, 144. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Zhang, J.; Zhang, Q.; Ding, X. Using different single-step strategies to improve the efficiency of genomic prediction on body measurement traits in Pig. Front. Genet. 2019, 9, 730. [Google Scholar] [CrossRef]
- Larsson, R.; Rydén, J. Applications of discrete factor analysis. Commun. Stat-Simul. C 2021, 50, 1–11. [Google Scholar] [CrossRef]
- Chu, M.; Shi, S. Phenotypic factor analysis for linear type traits in Beijing Holstein cows. Asian Australas. J. Anim. Sci. 2002, 15, 1527–1530. [Google Scholar] [CrossRef]
- Sieber, M.; Freeman, A.E.; Hinz, P.N. Factor analysis for evaluating relationships between first lactation type scores and production data of Holstein dairy cows. J. Dairy Sci. 1987, 70, 1018–1026. [Google Scholar] [CrossRef]
- Ali, A.K.; Koots, K.R.; Burnside, E.B. Factor analysis of genetic evaluations for type traits of Canadian Holstein sires and cows. Asian Australas. J. Anim. Sci. 1998, 11, 463–469. [Google Scholar] [CrossRef]
- Brown, J.E.; Brown, C.J.; Butts, W.T. Evaluating relationships among immature measures of size, shape and performance of beef bulls. I. Principal Components as Measures of Size and Shape in Young Hereford and Angus Bulls. J. Anim. Sci. 1973, 36, 1010–1020. [Google Scholar] [CrossRef]
- Kaiser, H. The varimax criterion for analytic rotation in factor analysis. Psychometrika 1958, 23, 187–200. [Google Scholar] [CrossRef]
- Mantovani, R.I.C.; Contiero, B. Factor analysis for genetic evaluation of linear type traits in dual purpose breeds. Ital. J. Anim. Sci. 2010, 4, 31–33. [Google Scholar] [CrossRef]
- Samoré, A.; Rizzi, R.; Rossoni, A.; Bagnato, A. Genetic parameters for functional longevity, type traits, SCS, milk flow and production in the Italian Brown Swiss. Ital. J. Anim. Sci. 2010, 9, 28. [Google Scholar] [CrossRef]
- Budianto, D.; Widi, T.S.; Panjono, P.; Budisatria, I.G.S.I.; Hartatik, T. Estimation of body weight using linear body measurements in two crossbred beef cattle. In Proceedings of the 9th International Seminar on Tropical Animal Production (ISTAP 2021), Yogyakarta, Indonesia, 21–22 September 2021; pp. 332–337. [Google Scholar]
- Manafiazar, G.; Goonewardene, L.; Miglior, F.; Crews, D.; Basarab, J.; Okine, E.; Wang, A. Genetic and phenotypic correlations among feed efficiency, production and selected conformation traits in dairy cows. Animal 2015, 10, 1–9. [Google Scholar] [CrossRef]
- Dadati, E.; Kennedy, B.; Burnside, E. Relationships between conformation and reproduction in Holstein cows: Type and calving performance. J. Dairy Sci. 1985, 68, 2639–2645. [Google Scholar] [CrossRef]
- Van, D.R.; Boettcher, P.; Schaeffer, L. Genetics of locomotion. Livest. Prod. Sci. 2004, 90, 247–253. [Google Scholar]
- Pérez-Cabal, M.; Charfeddine, N. Short communication: Association of foot and leg conformation and body weight with claw disorders in Spanish Holstein cows. J. Dairy Sci. 2016, 99, 9104–9108. [Google Scholar] [CrossRef]
- Rogers, G.W.; Banos, G.; Nielsen, U.; Philipsson, J. Genetic correlations among somatic cell scores, productive life, and type traits from the United States and udder health measures from Denmark and Sweden. J. Dairy Sci. 1998, 81, 1445–1453. [Google Scholar] [CrossRef]
- Chrystal, M.; Seykora, A.; Hansen, L.B. Heritabilities of teat end shape and teat diameter and their relationships with somatic cell score. J. Dairy Sci. 1999, 82, 2017–2022. [Google Scholar] [CrossRef]
- Němcová, E.; Štípková, M.; Zavadilová, L.; Bouška, J.; Vacek, M. The relationship between somatic cell count, milk production and six linearly scored type traits in Holstein cows. Czech. J. Anim. Sci. 2007, 52, 437–446. [Google Scholar]
- DeGroot, B.; Keown, J.; Van Vleck, L.; Marotz, E. Genetic parameters and responses of linear type, yield traits, and somatic cell scores to divergent selection for predicted transmitting ability for type in Holsteins1. J. Dairy Sci. 2002, 85, 1578–1585. [Google Scholar] [CrossRef]
- Dube, B.; Dzama, K.; Banga, C. Genetic analysis of somatic cell score and udder type traits in South African Holstein cows. S. Afr. J. Anim. Sci. 2008, 38, 1–11. [Google Scholar] [CrossRef] [Green Version]
| Traits | Number | Minimum | Maximum | Average | SD | CV (%) |
|---|---|---|---|---|---|---|
| Composite trait | ||||||
| Final score (points) | 1016 | 74.6 | 87.1 | 82.2 | 2.1 | 2.5 |
| Body frame (points) | 1016 | 68.5 | 95.0 | 85.1 | 4.6 | 5.4 |
| Muscularity (points) | 1016 | 70.5 | 91.5 | 80.7 | 3.1 | 3.9 |
| Rump (points) | 1016 | 58.0 | 95.0 | 80.4 | 6.2 | 7.7 |
| feet and legs (points) | 1016 | 73.0 | 94.3 | 86.7 | 3.6 | 4.2 |
| Mammary system (points) | 1016 | 64.2 | 88.1 | 78.8 | 4.2 | 5.4 |
| Individual body conformation trait | ||||||
| Body frame | ||||||
| Stature (cm) | 1016 | 126 | 154 | 140.7 | 4.5 | 3.2 |
| Body depth (cm) | 1016 | 61 | 90 | 78.4 | 6.0 | 7.6 |
| Chest width (cm) | 1016 | 18 | 39 | 27.6 | 4.3 | 15.6 |
| Muscularity | ||||||
| Withers width (cm) | 1016 | 10 | 28 | 18.4 | 2.9 | 15.6 |
| Hind leg half circumference (cm) | 1016 | 33 | 52 | 42.5 | 3.0 | 6.9 |
| Rear leg height (cm) | 1016 | 62 | 88 | 76.6 | 4.3 | 5.6 |
| Rib and bone (points) | 1016 | 1 | 9 | 6.8 | 1.2 | 18.0 |
| Rump | ||||||
| Rump length (cm) | 1016 | 43 | 59 | 51.8 | 3.1 | 5.9 |
| Rump width (cm) | 1016 | 17 | 29 | 22.7 | 1.9 | 8.4 |
| Rump angle (cm) | 1016 | −5 | 17 | 6.3 | 3.3 | 52.7 |
| Feet and legs | ||||||
| Heel depth (cm) | 1016 | 1 | 8 | 4.1 | 0.9 | 21.7 |
| Foot angle (points) | 1016 | 2 | 9 | 5.4 | 1.1 | 20.0 |
| Rear legs side view (points) | 1016 | 2 | 9 | 5.2 | 1.2 | 22.4 |
| Bone quality (points) | 1016 | 4 | 8 | 6.1 | 0.6 | 9.8 |
| Rear legs rear view (points) | 1016 | 2 | 9 | 5.2 | 1.2 | 22.4 |
| Mammary system | ||||||
| Rear Udder height (cm) | 1016 | 14 | 38 | 29.0 | 4.2 | 14.4 |
| Rear Udder width (cm) | 1016 | 7 | 24 | 13.0 | 2.9 | 22.4 |
| Median suspensory (cm) | 1016 | 0 | 8 | 3.5 | 1.5 | 42.9 |
| Udder depth (cm) | 1016 | −19 | 22 | 6.7 | 5.2 | 77.2 |
| Fore udder length (cm) | 1016 | 10 | 29 | 17.7 | 3.4 | 19.2 |
| Front teat length (cm) | 1016 | 2 | 10 | 4.8 | 1.3 | 27.2 |
| Front teat diameter (cm) | 1016 | 1 | 4 | 2.5 | 0.5 | 21.3 |
| Fore udder attachment (points) | 1016 | 1 | 8 | 4.2 | 1.3 | 32.0 |
| Rear udder length (points) | 1016 | 1 | 9 | 5.0 | 1.5 | 29.7 |
| Udder balance (points) | 1016 | 1 | 9 | 4.7 | 0.9 | 19.0 |
| Fore teat placement (points) | 1016 | 1 | 8 | 4.0 | 1.1 | 26.4 |
| Rear teat placement (points) | 1016 | 1 | 9 | 5.2 | 1.0 | 19.5 |
| A Matrix | H Matrix | |||||||
|---|---|---|---|---|---|---|---|---|
| Traits 1 | ||||||||
| Composite trait | ||||||||
| Final score | 0.72 | 3.33 | 4.05 | 0.18 ± 0.08 | 0.55 | 3.50 | 4.05 | 0.14 ± 0.07 |
| Body frame | 8.47 | 11.27 | 19.74 | 0.43 ± 0.13 | 9.36 | 10.48 | 19.84 | 0.47 ± 0.12 |
| Muscularity | 0.62 | 8.86 | 9.48 | 0.07 ± 0.05 | 0.99 | 8.53 | 9.52 | 0.10 ± 0.06 |
| Rump | 7.57 | 23.94 | 31.51 | 0.24 ± 0.10 | 6.65 | 24.74 | 31.39 | 0.21 ± 0.09 |
| Feet and leg | 1.29 | 11.37 | 12.66 | 0.10 ± 0.06 | 1.27 | 11.41 | 12.68 | 0.10 ± 0.06 |
| Mammary system | 2.74 | 10.63 | 13.37 | 0.20 ± 0.09 | 3.11 | 10.33 | 13.44 | 0.23 ± 0.09 |
| Average | 3.57 | 11.57 | 15.14 | 0.20 ± 0.09 | 3.66 | 11.50 | 15.15 | 0.21 ± 0.08 |
| Individual body conformation | ||||||||
| Body frame | ||||||||
| ST | 11.38 | 8.93 | 20.31 | 0.56 ± 0.12 | 13.48 | 7.17 | 20.66 | 0.65 ± 0.11 |
| BD | 5.27 | 25.33 | 30.60 | 0.17 ± 0.08 | 6.00 | 24.70 | 30.70 | 0.20 ± 0.09 |
| CW | 2.29 | 15.72 | 18.01 | 0.13 ± 0.08 | 2.04 | 15.96 | 18.00 | 0.11 ± 0.07 |
| Average | 6.31 | 16.66 | 22.97 | 0.29 ± 0.09 | 7.17 | 15.94 | 23.12 | 0.32 ± 0.09 |
| Muscularity | ||||||||
| WW | 0.77 | 7.51 | 8.28 | 0.09 ± 0.07 | 0.53 | 7.73 | 8.26 | 0.06 ± 0.06 |
| HLHC | 0.53 | 7.55 | 8.08 | 0.07 ± 0.05 | 0.64 | 7.45 | 8.09 | 0.08 ± 0.05 |
| RLH | 0.86 | 14.95 | 15.81 | 0.05 ± 0.05 | 0.94 | 14.88 | 15.82 | 0.06 ± 0.05 |
| RAB | 0.07 | 1.29 | 1.36 | 0.05 ± 0.05 | 0.52 | 1.31 | 1.83 | 0.04 ± 0.04 |
| Average | 0.56 | 7.83 | 8.38 | 0.07 ± 0.06 | 0.66 | 7.84 | 8.50 | 0.06 ± 0.05 |
| Rump | ||||||||
| RL | 2.24 | 5.57 | 7.81 | 0.29 ± 0.11 | 2.67 | 5.23 | 7.90 | 0.34 ± 0.12 |
| RW | 0.68 | 2.40 | 3.08 | 0.22 ± 0.09 | 0.62 | 2.46 | 3.08 | 0.20 ± 0.08 |
| RA | 1.39 | 7.85 | 9.24 | 0.15 ± 0.07 | 1.94 | 7.35 | 9.29 | 0.21 ± 0.08 |
| Average | 1.44 | 5.27 | 6.71 | 0.22 ± 0.09 | 1.74 | 5.01 | 6.76 | 0.25 ± 0.09 |
| Feet and legs | ||||||||
| HD | 0.03 | 0.59 | 0.62 | 0.05 ± 0.05 | 0.05 | 0.58 | 0.63 | 0.07 ± 0.05 |
| FA | 0.11 | 0.83 | 0.94 | 0.11 ± 0.06 | 0.15 | 0.78 | 0.93 | 0.16 ± 0.07 |
| RLSV | 0.11 | 1.18 | 1.29 | 0.09 ± 0.06 | 0.09 | 1.20 | 1.29 | 0.07 ± 0.05 |
| BQ | 0.02 | 0.32 | 0.34 | 0.07 ± 0.05 | 0.04 | 0.30 | 0.34 | 0.12 ± 0.06 |
| RLRV | 0.19 | 1.34 | 1.53 | 0.12 ± 0.07 | 0.20 | 1.33 | 1.53 | 0.13 ± 0.07 |
| Average | 0.09 | 0.85 | 0.94 | 0.09 ± 0.06 | 0.11 | 0.84 | 0.94 | 0.11 ± 0.06 |
| Mammary system | ||||||||
| RUH | 3.08 | 14.72 | 17.80 | 0.17 ± 0.07 | 3.00 | 14.82 | 17.82 | 0.17 ± 0.07 |
| RUW | 0.57 | 7.06 | 7.63 | 0.07 ± 0.05 | 0.83 | 6.81 | 7.64 | 0.11 ± 0.06 |
| MS | 0.11 | 1.73 | 1.84 | 0.06 ± 0.05 | 0.18 | 1.67 | 1.85 | 0.10 ± 0.06 |
| UD | 4.01 | 14.15 | 18.16 | 0.22 ± 0.09 | 4.71 | 13.55 | 18.26 | 0.26 ± 0.09 |
| FUL | 1.02 | 7.91 | 8.93 | 0.11 ± 0.06 | 1.08 | 7.87 | 8.95 | 0.12 ± 0.07 |
| FTL | 0.21 | 1.57 | 1.78 | 0.12 ± 0.06 | 0.20 | 1.58 | 1.78 | 0.11 ± 0.06 |
| FTD | 0.05 | 0.23 | 0.28 | 0.18 ± 0.07 | 0.05 | 0.23 | 0.28 | 0.18 ± 0.07 |
| FUA | 0.31 | 1.34 | 1.65 | 0.19 ± 0.08 | 0.44 | 1.23 | 1.67 | 0.27 ± 0.09 |
| RUL | 0.14 | 1.75 | 1.89 | 0.07 ± 0.06 | 0.05 | 1.83 | 1.88 | 0.03 ± 0.05 |
| UB | 0.22 | 0.61 | 0.83 | 0.26 ± 0.10 | 0.28 | 0.54 | 0.82 | 0.34 ± 0.09 |
| FTP | 0.19 | 0.78 | 0.97 | 0.20 ± 0.08 | 0.20 | 0.78 | 0.98 | 0.20 ± 0.08 |
| RTP | 0.16 | 0.76 | 0.92 | 0.17 ± 0.08 | 0.20 | 0.72 | 0.92 | 0.22 ± 0.09 |
| Average | 0.84 | 4.38 | 5.22 | 0.15 ± 0.07 | 0.94 | 4.30 | 5.24 | 0.18 ± 0.07 |
| Factor | Eigenvalue | Proportional Variance (%) | Cumulative Variance (%) |
|---|---|---|---|
| F1 | 3.65 | 13.51 | 13.51 |
| F2 | 2.20 | 8.13 | 21.65 |
| F3 | 1.98 | 7.32 | 28.96 |
| F4 | 1.62 | 6.01 | 34.97 |
| F5 | 1.40 | 5.18 | 40.15 |
| F6 | 1.33 | 4.93 | 45.08 |
| F7 | 1.21 | 4.49 | 49.57 |
| F8 | 1.13 | 4.18 | 53.75 |
| F9 | 1.03 | 3.80 | 57.55 |
| F10 | 0.96 | 3.56 | 61.12 |
| F11 | 0.93 | 3.46 | 64.58 |
| F12 | 0.89 | 3.30 | 67.88 |
| F13 | 0.86 | 3.18 | 71.05 |
| F14 | 0.81 | 3.01 | 74.06 |
| F15 | 0.74 | 2.74 | 76.81 |
| F16 | 0.73 | 2.73 | 79.54 |
| F17 | 0.71 | 2.65 | 82.19 |
| F18 | 0.67 | 2.47 | 84.66 |
| F19 | 0.62 | 2.29 | 86.94 |
| F20 | 0.59 | 2.17 | 89.11 |
| F21 | 0.56 | 2.09 | 91.20 |
| F22 | 0.49 | 1.81 | 93.00 |
| F23 | 0.43 | 1.60 | 94.60 |
| F24 | 0.41 | 1.52 | 96.13 |
| Varimax Latent Factors | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Trait 1 | F1 | F2 | F3 | F4 | F5 | F6 | F7 | F8 | F9 | Communality |
| ST | 0.65 | 0.68 | ||||||||
| BD | 0.46 | −0.51 | 0.68 | |||||||
| CW | −0.45 | 0.39 | ||||||||
| WW | −0.44 | 0.54 | ||||||||
| HLHC | 0.60 | 0.43 | ||||||||
| RLH | 0.46 | 0.54 | ||||||||
| RAB | −0.42 | 0.44 | ||||||||
| RL | 0.72 | 0.62 | ||||||||
| RW | 0.57 | 0.49 | ||||||||
| RA | 0.83 | 0.72 | ||||||||
| HD | 0.76 | 0.64 | ||||||||
| FA | 0.77 | 0.67 | ||||||||
| RLSV | 0.44 | |||||||||
| BQ | 0.46 | |||||||||
| RLRV | 0.79 | 0.68 | ||||||||
| RAH | −0.53 | 0.52 | ||||||||
| RUW | 0.58 | 0.60 | ||||||||
| MS | 0.66 | 0.51 | ||||||||
| UD | 0.50 | 0.63 | ||||||||
| FUL | 0.70 | 0.60 | ||||||||
| FTL | 0.80 | 0.68 | ||||||||
| FTD | 0.81 | 0.72 | ||||||||
| FUA | 0.53 | 0.51 | ||||||||
| RUL | 0.50 | 0.55 | ||||||||
| UB | 0.65 | 0.48 | ||||||||
| FTP | 0.79 | 0.63 | ||||||||
| RTP | 0.73 | 0.68 | ||||||||
| Variance explained (%) | 2.32 | 1.98 | 1.91 | 1.85 | 1.69 | 1.68 | 1.52 | 1.33 | 1.26 | |
| A Matrix | H Matrix | |||||||
|---|---|---|---|---|---|---|---|---|
| Factor Score | ||||||||
| F1 | 0.39 | 0.52 | 0.91 | 0.43 ± 0.13 | 0.39 | 0.52 | 0.91 | 0.43 ± 0.12 |
| F2 | 0.04 | 0.64 | 0.68 | 0.06 ± 0.05 | 0.07 | 0.62 | 0.69 | 0.10 ± 0.06 |
| F3 | 0.16 | 0.61 | 0.77 | 0.21 ± 0.08 | 0.16 | 0.62 | 0.78 | 0.21 ± 0.09 |
| F4 | 0.008 | 0.92 | 0.93 | 0.008 ± 0.04 | 0.04 | 0.90 | 0.94 | 0.04 ± 0.05 |
| F5 | 0.20 | 0.71 | 0.91 | 0.22 ± 0.10 | 0.24 | 0.68 | 0.92 | 0.26 ± 0.11 |
| F6 | 0.09 | 0.70 | 0.79 | 0.11 ± 0.07 | 0.11 | 0.68 | 0.79 | 0.14 ± 0.08 |
| F7 | 0.06 | 0.60 | 0.66 | 0.09 ± 0.05 | 0.07 | 0.59 | 0.66 | 0.10 ± 0.06 |
| F8 | 0.08 | 0.58 | 0.66 | 0.12 ± 0.06 | 0.10 | 0.57 | 0.67 | 0.15 ± 0.07 |
| F9 | 0.31 | 0.58 | 0.89 | 0.35 ± 0.12 | 0.34 | 0.55 | 0.89 | 0.38 ± 0.12 |
| Composite Traits | F1 | F2 | F3 | F4 | F5 | F6 | F7 | F8 | F9 |
|---|---|---|---|---|---|---|---|---|---|
| Body frame | 0.67 | −0.31 | 0.28 | ||||||
| Muscularity | 0.55 | 0.24 | 0.23 | 0.20 | −0.24 | 0.45 | |||
| Rump | 0.58 | 0.24 | 0.23 | 0.44 | |||||
| feet and legs | 0.20 | ||||||||
| Mammary system | 0.30 | 0.31 | 0.44 |
| Composite Traits | F1 | F2 | F3 | F4 | F5 | F6 | F7 | F8 | F9 |
|---|---|---|---|---|---|---|---|---|---|
| Body frame | 0.69 | −0.34 | 0.36 | ||||||
| Muscularity | 0.59 | 0.31 | 0.23 | 0.22 | −0.25 | 0.58 | |||
| Rump | 0.64 | 0.33 | 0.28 | 0.51 | |||||
| feet and legs | 0.33 | ||||||||
| Mammary system | 0.39 | 0.32 | 0.50 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, L.; Luo, H.; Zhang, X.; Lu, H.; Zhang, M.; Ge, J.; Zhang, T.; Yan, M.; Tan, X.; Huang, X.; et al. Factor Analysis of Genetic Parameters for Body Conformation Traits in Dual-Purpose Simmental Cattle. Animals 2022, 12, 2433. https://doi.org/10.3390/ani12182433
Xu L, Luo H, Zhang X, Lu H, Zhang M, Ge J, Zhang T, Yan M, Tan X, Huang X, et al. Factor Analysis of Genetic Parameters for Body Conformation Traits in Dual-Purpose Simmental Cattle. Animals. 2022; 12(18):2433. https://doi.org/10.3390/ani12182433
Chicago/Turabian StyleXu, Lei, Hanpeng Luo, Xiaoxue Zhang, Haibo Lu, Menghua Zhang, Jianjun Ge, Tao Zhang, Mengjie Yan, Xueting Tan, Xixia Huang, and et al. 2022. "Factor Analysis of Genetic Parameters for Body Conformation Traits in Dual-Purpose Simmental Cattle" Animals 12, no. 18: 2433. https://doi.org/10.3390/ani12182433






