High-Energy Supplemental Feeding Shifts Gut Microbiota Composition and Function in Red Deer (Cervus elaphus)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Fecal Sample Collection
2.3. DNA Extraction and PCR Amplification
2.4. Library Construction and Sequencing
2.5. Sequence Processing and Analysis
2.6. Alpha and Beta-Diversity Analyses
2.7. Analysis of Species Composition and Function
3. Results
3.1. Sequencing Information
3.2. Analysis of Gut Microbiota Diversity
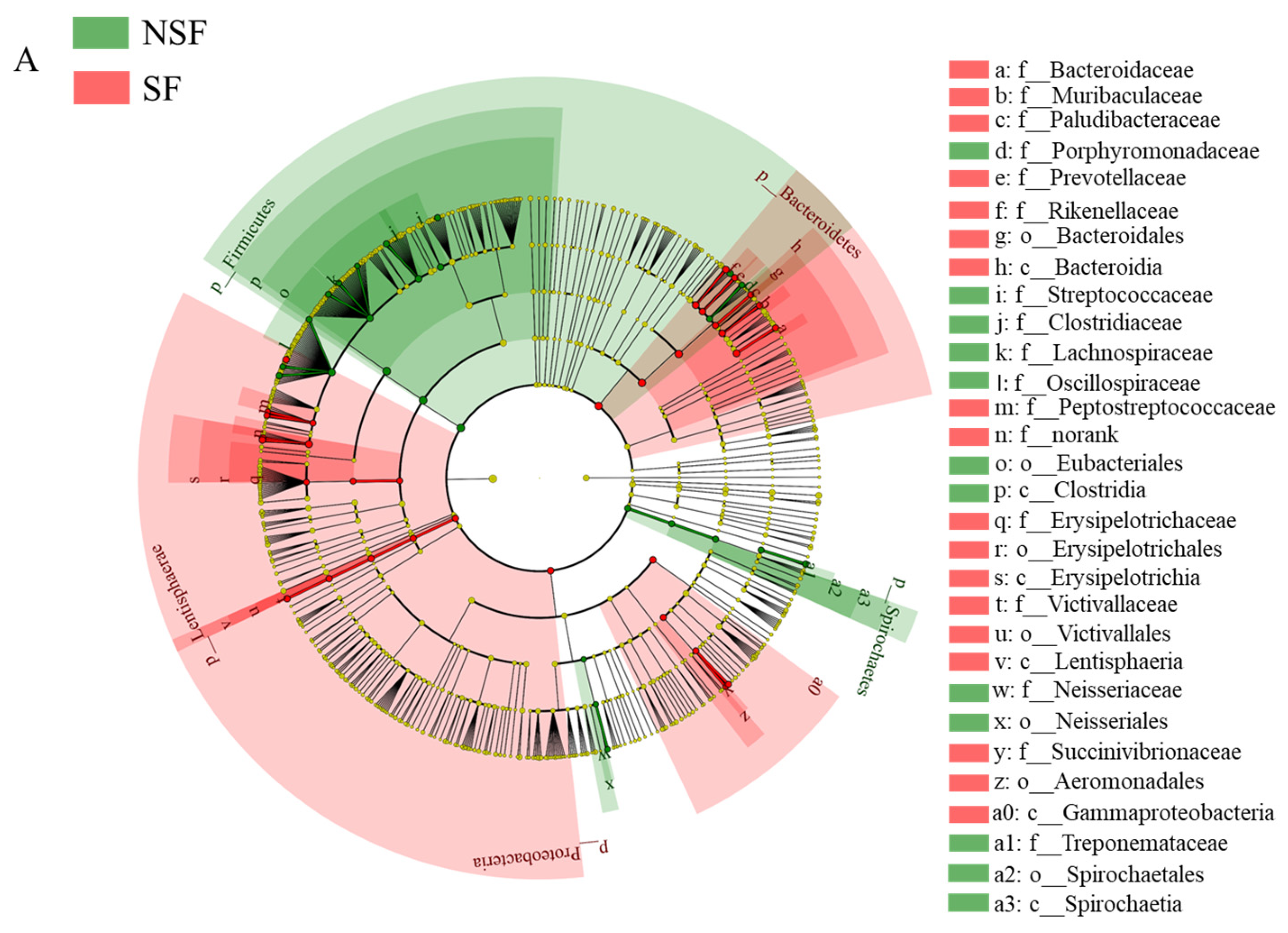
3.3. Gut Microbiota Community Composition
3.4. Functional Analysis of Gut Microbiota in Red Deer
4. Discussion
4.1. Presumption 1
4.2. Presumption 2
4.3. Presumption 3
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dearing, M.D.; Kohl, K.D. Beyond fermentation: Other important services provided to endothermic herbivores by their gut microbiota. Integr. Comp. Biol. 2017, 57, 723–731. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Tu, Y.; Sun, J.; Cai, P.; Zhou, Y.; Huang, Y.; Zhang, S.; Chen, W.; Wang, L.; Du, M. Fermented mixed feed regulates intestinal microbial community and metabolism and alters pork flavor and umami. Meat Sci. 2023, 201, 109177. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.; Webb, M.; Ghimire, S.; Blair, A.; Olson, K.; Fenske, G.J.; Fonder, A.T.; Christopher-Hennings, J.; Brake, D.; Scaria, J. Metagenomic characterization of the effect of feed additives on the gut microbiome and antibiotic resistome of feedlot cattle. Sci. Rep. 2017, 7, 12257. [Google Scholar] [CrossRef] [PubMed]
- Cortes-Ortiz, L.; Amato, K.R. Host genetics influence the gut microbiome. Science 2021, 373, 159–160. [Google Scholar] [CrossRef] [PubMed]
- Mizukami, K.; Uchiyama, J.; Igarashi, H.; Murakami, H.; Osumi, T.; Shima, A.; Ishiahra, G.; Nasukawa, T.; Une, Y.; Sakaguchi, M. Age-related analysis of the gut microbiome in a purebred dog colony. FEMS Microbiol. Lett. 2019, 366, 095. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Xie, H.; Yang, Z.; Chen, J.; Yin, J.; Tian, P.; Wang, H.; Zhao, J.; Zhang, H.; Lu, W. Statistical modeling of gut microbiota for personalized health status monitoring. Microbiome 2023, 11, 184. [Google Scholar] [CrossRef] [PubMed]
- Ilina, L.A.; Filippova, V.A.; Brazhnik, E.A.; Dubrovin, A.V.; Yildirim, E.A.; Dunyashev, T.P.; Laptev, G.Y.; Novikova, N.I.; Sobolev, D.V.; Yuzhakov, A.A. The comparative analysis of the ruminal bacterial population in Reindeer (Rangifer tarandus L.) from the Russian Arctic Zone: Regional and seasonal effects. Animals 2021, 11, 911. [Google Scholar] [CrossRef]
- Qin, W.; Song, P.; Zhang, S. Seasonal and soil microbiota effects on the adaptive strategies of wild Goitered Gazelles based on the gut microbiota. Front. Microbiol. 2022, 13, 918090. [Google Scholar] [CrossRef]
- Kim, P.; Shin, N.; Lee, J.; Kim, M.; Whon, T.; Hyun, D.; Yun, J.; Jung, M.; Kim, J.; Bae, J. Host habitat is the major determinant of the gut microbiome of fish. Microbiome 2021, 9, 166. [Google Scholar] [CrossRef]
- Guo, W.; Mishra, S.; Wang, C.; Zhang, H.; Ning, R.; Kong, F.; Zeng, B.; Zhao, J.; Li, Y. Comparative study of gut microbiota in wild and captive giant pandas (Ailuropoda melanoleuca). Genes 2019, 10, 827. [Google Scholar] [CrossRef]
- Xi, L.; Han, J.; Wen, X.; Zhao, L.; Qin, X.; Luo, S.; Lv, D.; Song, S. Species variations in the gut microbiota of captive snub-nosed monkeys. Front. Endocrinol. 2023, 14, 1250865. [Google Scholar] [CrossRef] [PubMed]
- Gibson, K.M.; Nguyen, B.N.; Neumann, L.M.; Miller, M.; Buss, P.; Daniels, S.; Ahn, M.J.; Crandall, K.A.; Pukazhenthi, B. Gut microbiome differences between wild and captive black rhinoceros–implications for rhino health. Sci. Rep. 2019, 9, 7570. [Google Scholar] [CrossRef] [PubMed]
- Pacheco-Torres, I.; Hernández-Sánchez, D.; García-De la Peña, C.; Tarango-Arámbula, L.A.; Crosby-Galván, M.M.; Sánchez-Santillán, P. Analysis of the intestinal and faecal bacterial microbiota of the Cervidae family using 16S next-generation sequencing: A review. Microorganisms 2023, 11, 1860. [Google Scholar] [CrossRef] [PubMed]
- Lovari, S.; Lorenzini, R.; Masseti, M.; Pereladova, O.; Carden, R.; Brook, S.; Mattioli, S. Cervus elaphus (Errata Version Published in 2019). The IUCN Red List of Threatened Species 2018: E. T55997072A142404453. Available online: https://www.iucnredlist.org/species/55997072/142404453 (accessed on 16 March 2020).
- Wolf, J.F.; Kriss, K.D.; MacAulay, K.M.; Munro, K.; Patterson, B.R.; Shafer, A.B. Gut microbiome composition predicts summer core range size in two divergent ungulates. FEMS Microbiol. Ecol. 2021, 97, fiab048. [Google Scholar] [CrossRef]
- You, Z.; Deng, J.; Liu, J.; Fu, J.; Xiong, H.; Luo, W.; Xiong, J. Seasonal variations in the composition and diversity of gut microbiota in white-lipped deer (Cervus albirostris). PeerJ 2022, 10, e13753. [Google Scholar] [CrossRef]
- Eddington, H.S.; Carroll, C.; Larsen, R.T.; McMillan, B.R.; Chaston, J.M. Spatiotemporal variation in the fecal microbiota of mule deer is associated with proximate and future measures of host health. BMC Vet. Res. 2021, 17, 258. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, G.T.; Craine, J.M.; Robeson, M.S.; Fierer, N. Seasonal shifts in diet and gut microbiota of the American bison (Bison bison). PLoS ONE 2015, 10, e0142409. [Google Scholar] [CrossRef] [PubMed]
- Maurice, C.F.; Knowles, S.C.L.; Ladau, J.; Pollard, K.S.; Fenton, A.; Pedersen, A.B.; Turnbaugh, P.J. Marked seasonal variation in the wild mouse gut microbiota. ISME J. 2015, 9, 2423–2434. [Google Scholar] [CrossRef] [PubMed]
- Baniel, A.; Amato, K.R.; Beehner, J.C.; Bergman, T.J.; Mercer, A.; Perlman, R.F.; Petrullo, L.; Reitsema, L.; Sams, S.; Lu, A. Seasonal shifts in the gut microbiome indicate plastic responses to diet in wild geladas. Microbiome 2021, 9, 26. [Google Scholar] [CrossRef]
- Couch, C.E.; Wise, B.L.; Scurlock, B.M.; Rogerson, J.D.; Fuda, R.K.; Cole, E.K.; Szcodronski, K.E.; Sepulveda, A.J.; Hutchins, P.R.; Cross, P.C. Effects of supplemental feeding on the fecal bacterial communities of Rocky Mountain elk in the Greater Yellowstone Ecosystem. PLoS ONE 2021, 16, e0249521. [Google Scholar] [CrossRef]
- Zhen, J.; Ren, Y.; Zhang, H.; Yuan, X.; Wang, L.; Shen, H.; Liu, P.; Chen, Y. Effect of different dietary regimes on the gut microbiota and fecal metabolites of Pere David’s deer. Animals 2022, 12, 584. [Google Scholar] [CrossRef] [PubMed]
- Menke, S.; Heurich, M.; Henrich, M.; Wilhelm, K.; Sommer, S. Impact of winter enclosures on the gut bacterial microbiota of red deer in the Bavarian Forest National Park. Wildl. Biol. 2019, 2019, 1–10. [Google Scholar] [CrossRef]
- Sun, Y.; Yu, Y.; Guo, J.; Zhong, L.; Zhang, M. Alterations in fecal microbiota linked to environment and sex in red deer (Cervus elaphus). Animals 2023, 13, 929. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Yu, Y.; Guo, J.; Zhong, L.; Zhang, M. Comparative Analysis of Gut Microbiome between Captive and Wild Red Deer 1 (Cervus elaphus) in Inner Mongolia. 2021. Available online: https://www.researchsquare.com/article/rs-312376/v1 (accessed on 10 January 2024).
- Westoby, M. An analysis of diet selection by large generalist herbivores. Am. Nat. 1974, 108, 290–304. [Google Scholar] [CrossRef]
- Owen-Smith, N.; Novellie, P. What should a clever ungulate eat? Am. Nat. 1982, 119, 151–178. [Google Scholar] [CrossRef]
- Belovsky, G.E. Optimal foraging and community structure: Implications for a guild of generalist grassland herbivores. Oecologia 1986, 70, 35–52. [Google Scholar] [CrossRef] [PubMed]
- Schoener, T.W. Theory of feeding strategies. Annu. Rev. Ecol. Syst. 1971, 2, 369–404. [Google Scholar] [CrossRef]
- Hixon, M.A. Energy maximizers and time minimizers: Theory and reality. Am. Nat. 1982, 119, 596–599. [Google Scholar] [CrossRef]
- Ward, D. The role of satisficing in foraging theory. Oikos 1992, 63, 312. [Google Scholar] [CrossRef]
- Van Wieren, S. Do large herbivores select a diet that maximizes short-term energy intake rate? For. Ecol. Manag. 1996, 88, 149–156. [Google Scholar] [CrossRef]
- Ricci, S.; Sandfort, R.; Pinior, B.; Mann, E.; Wetzels, S.U.; Stalder, G. Impact of supplemental winter feeding on ruminal microbiota of roe deer Capreolus capreolus. Wildl. Biol. 2019, 2019, 1–11. [Google Scholar] [CrossRef]
- Mason, F.; Fotschki, B.; Di Rosso, A.; Korzekwa, A. Influence of farming conditions on the rumen of red deer (Cervus elaphus). Animals 2019, 9, 601. [Google Scholar] [CrossRef]
- Fang, J. Natural vegetation restoration and ecological gardening in wildlife parks. For. Xinjiang 2010, 5, 37–39. [Google Scholar]
- Ando, H.; Mukai, H.; Komura, T.; Dewi, T.; Ando, M.; Isagi, Y. Methodological trends and perspectives of animal dietary studies by noninvasive fecal DNA metabarcoding. Environ. DNA 2020, 2, 391–406. [Google Scholar] [CrossRef]
- Zhou, C.L.; Muhammat, A.; Turdy, R.; Halik, M. Sex ratio and winter home range of Tianshan red deer (Cervus elaphus songaricus) by noninvasive method. Sci. Technol. Rev. 2015, 33, 91–96. [Google Scholar]
- Li, J.L. Intestinal Flora Structure of Red Deer (Cervus elaphus) in Winter and Summer; Northeast Agricultural University: Harbin, China, 2020. [Google Scholar]
- Tende, T.; Hansson, B.; Ottosson, U.; Bensch, S. Evaluating preservation medium for the storage of DNA in African lion Panthera leo faecal samples. Curr. Zool. 2014, 60, 351–358. [Google Scholar] [CrossRef]
- Guo, N.; Wu, Q.; Shi, F.; Niu, J.; Zhang, T.; Degen, A.A.; Fang, Q.; Ding, L.; Shang, Z.; Zhang, Z. Seasonal dynamics of diet–gut microbiota interaction in adaptation of yaks to life at high altitude. npj Biofilms Microbiomes 2021, 7, 38. [Google Scholar] [CrossRef]
- Notario, E.; Visci, G.; Fosso, B.; Gissi, C.; Tanaskovic, N.; Rescigno, M.; Marzano, M.; Pesole, G. Amplicon-based microbiome profiling: From second-to third-generation sequencing for higher taxonomic resolution. Genes 2023, 14, 1567. [Google Scholar] [CrossRef]
- Kim, M.; Chun, J. 16S rRNA gene-based identification of bacteria and archaea using the EzTaxon server. Methods Microbiol. 2014, 41, 61–74. [Google Scholar]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Lozupone, C.; Lladser, M.E.; Knights, D.; Stombaugh, J.; Knight, R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011, 5, 169–172. [Google Scholar] [CrossRef]
- Anderson, M.J.; Walsh, D.C. PERMANOVA, ANOSIM, and the Mantel test in the face of heterogeneous dispersions: What null hypothesis are you testing? Ecol. Monogr. 2013, 83, 557–574. [Google Scholar] [CrossRef]
- Xiong, W.; Song, Y.; Yang, K.; Gu, Y.; Wei, Z.; Kowalchuk, G.A.; Xu, Y.; Jousset, A.; Shen, Q.; Geisen, S. Rhizosphere protists are key determinants of plant health. Microbiome 2020, 8, 27. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B-Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Dubé, C.; Ziegler, M.; Mercière, A.; Boissin, E.; Planes, S.; Bourmaud, C.F.; Voolstra, C.R. Naturally occurring fire coral clones demonstrate a genetic and environmental basis of microbiome composition. Nat. Commun. 2021, 12, 6402. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Ijaz, M.U.; Ahmed, M.I.; Zou, X.; Hussain, M.; Zhang, M.; Zhao, F.; Xu, X.; Zhou, G.; Li, C. Beef, casein, and soy proteins differentially affect lipid metabolism, triglycerides accumulation and gut microbiota of high-fat diet-fed C57BL/6J mice. Front. Microbiol. 2018, 9, 2200. [Google Scholar] [CrossRef]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef]
- McKenzie, V.J.; Song, S.J.; Delsuc, F.; Prest, T.L.; Oliverio, A.M.; Korpita, T.M.; Alexiev, A.; Amato, K.R.; Metcalf, J.L.; Kowalewski, M. The effects of captivity on the mammalian gut microbiome. Integr. Comp. Biol. 2017, 57, 690–704. [Google Scholar] [CrossRef]
- Sun, C.H.; Liu, H.Y.; Liu, B.; Yuan, B.D.; Lu, C.H. Analysis of the gut microbiome of wild and captive Père David’s deer. Front. Microbiol. 2019, 10, 2331. [Google Scholar] [CrossRef]
- Kartzinel, T.R.; Hsing, J.C.; Musili, P.M.; Brown, B.R.; Pringle, R.M. Covariation of diet and gut microbiome in African megafauna. Proc. Natl. Acad. Sci. USA 2019, 116, 23588–23593. [Google Scholar] [CrossRef]
- Minich, D.; Madden, C.; Evans, M.V.; Ballash, G.A.; Barr, D.J.; Poulsen, K.P.; Dennis, P.M.; Hale, V.L. Alterations in gut microbiota linked to provenance, sex, and chronic wasting disease in white-tailed deer (Odocoileus virginianus). Sci. Rep. 2021, 11, 13218. [Google Scholar] [CrossRef]
- Wen, Y.; Li, S.; Wang, Z.; Feng, H.; Yao, X.; Liu, M.; Chang, J.; Ding, X.; Zhao, H.; Ma, W. Intestinal microbial diversity of free-range and captive yak in Qinghai Province. Microorganisms 2022, 10, 754. [Google Scholar] [CrossRef]
- Jiang, F.; Gao, H.; Qin, W.; Song, P.; Wang, H.; Zhang, J.; Liu, D.; Wang, D.; Zhang, T. Marked seasonal variation in structure and function of gut microbiota in forest and alpine musk deer. Front. Microbiol. 2021, 12, 699797. [Google Scholar] [CrossRef]
- Sorbara, M.T.; Littmann, E.R.; Fontana, E.; Moody, T.U.; Kohout, C.E.; Gjonbalaj, M.; Eaton, V.; Seok, R.; Leiner, I.M.; Pamer, E.G. Functional and genomic variation between human-derived isolates of Lachnospiraceae reveals inter-and intra-species diversity. Cell Host Microbe 2020, 28, 134–146.e4. [Google Scholar] [CrossRef]
- Sabathé, F.; Bélaïch, A.; Soucaille, P. Characterization of the cellulolytic complex (cellulosome) of Clostridium acetobutylicum. FEMS Microbiol. Lett. 2002, 217, 15–22. [Google Scholar] [CrossRef]
- Larue, R.; Yu, Z.; Parisi, V.A.; Egan, A.R.; Morrison, M. Novel microbial diversity adherent to plant biomass in the herbivore gastrointestinal tract, as revealed by ribosomal intergenic spacer analysis and rrs gene sequencing. Environ. Microbiol. 2005, 7, 530–543. [Google Scholar] [CrossRef]
- Lopetuso, L.R.; Scaldaferri, F.; Petito, V.; Gasbarrini, A. Commensal Clostridia: Leading players in the maintenance of gut homeostasis. Gut Pathog. 2013, 5, 23. [Google Scholar] [CrossRef]
- Bui, T.P.N.; Troise, A.D.; Nijsse, B.; Roviello, G.N.; Fogliano, V.; de Vos, W.M. Intestinimonas-like bacteria are important butyrate producers that utilize Nε-fructosyllysine and lysine in formula-fed infants and adults. J. Funct. Foods 2020, 70, 103974. [Google Scholar] [CrossRef]
- Thingholm, L.B.; Rühlemann, M.C.; Koch, M.; Fuqua, B.; Laucke, G.; Boehm, R.; Bang, C.; Franzosa, E.A.; Hübenthal, M.; Rahnavard, A. Obese individuals with and without type 2 diabetes show different gut microbial functional capacity and composition. Cell Host Microbe 2019, 26, 252–264.e10. [Google Scholar] [CrossRef]
- Bessa, R.J.; Alves, S.P.; Jerónimo, E.; Alfaia, C.M.; Prates, J.A.; Santos-Silva, J. Effect of lipid supplements on ruminal biohydrogenation intermediates and muscle fatty acids in lambs. Eur. J. Lipid Sci. Technol. 2007, 109, 868–878. [Google Scholar] [CrossRef]
- Lee, J.; Kim, H.J.; Lee, S.S.; Kim, K.W.; Kim, D.K.; Lee, S.H.; Lee, E.D.; Choi, B.H.; Barido, F.H.; Jang, A. Effects of diet and castration on fatty acid composition and volatile compounds in the meat of Korean native black goats. Anim. Biosci. 2023, 36, 962. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, M.; Iino, T.; Yuki, M.; Ohkuma, M. Lawsonibacter asaccharolyticus gen. nov., sp. nov., a butyrate-producing bacterium isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2018, 68, 2074–2081. [Google Scholar] [CrossRef]
- McIntyre, A.; Gibson, P.; Young, G. Butyrate production from dietary fibre and protection against large bowel cancer in a rat model. Gut 1993, 34, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Archer, S.Y.; Meng, S.; Shei, A.; Hodin, R.A. p21WAF1 is required for butyrate-mediated growth inhibition of human colon cancer cells. Proc. Natl. Acad. Sci. USA 1998, 95, 6791–6796. [Google Scholar] [CrossRef] [PubMed]
- Cox, L.M.; Yamanishi, S.; Sohn, J.; Alekseyenko, A.V.; Leung, J.M.; Cho, I.; Kim, S.G.; Li, H.; Gao, Z.; Mahana, D. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 2014, 158, 705–721. [Google Scholar] [CrossRef]
- Liao, Y.; Peng, Z.; Chen, L.; Nüssler, A.K.; Liu, L.; Yang, W. Deoxynivalenol, gut microbiota and immunotoxicity: A potential approach? Food Chem. Toxicol. 2018, 112, 342–354. [Google Scholar] [CrossRef]
- Zhang, Y.; Xing, H.; Bolotnikov, G.; Krämer, M.; Gotzmann, N.; Knippschild, U.; Kissmann, A.K.; Rosenau, F. Enriched aptamer libraries in fluorescence-based assays for Rikenella microfusus-specific gut microbiome analyses. Microorganisms 2023, 11, 2266. [Google Scholar] [CrossRef]
- Gnat, S.; Trościańczyk, A.; Nowakiewicz, A.; Majer-Dziedzic, B.; Ziółkowska, G.; Dziedzic, R.; Zięba, P.; Teodorowski, O. Experimental studies of microbial populations and incidence of zoonotic pathogens in the faeces of red deer (Cervus elaphus). Lett. Appl. Microbiol. 2015, 61, 446–452. [Google Scholar] [CrossRef]
- Huang, Y.; Ying, N.; Zhao, Q.; Chen, J.; Teow, S.Y.; Dong, W.; Lin, M.; Jiang, L.; Zheng, H. Amelioration of obesity-related disorders in high-fat diet-fed mice following fecal microbiota transplantation from Inulin-dosed mice. Molecules 2023, 28, 3997. [Google Scholar] [CrossRef] [PubMed]
- Zong, X.; Zhang, H.; Zhu, L.; Deehan, E.C.; Fu, J.; Wang, Y.; Jin, M. Auricularia auricula polysaccharides attenuate obesity in mice through gut commensal Papillibacter cinnamivorans. J. Adv. Res. 2023, 52, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Ge, T.; Yang, C.; Li, B.; Huang, X.; Zhao, L.; Zhang, X.; Tian, L.; Zhang, E. High-energy diet modify rumen microbial composition and microbial energy metabolism pattern in fattening sheep. BMC Vet. Res. 2023, 19, 32. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Xiao, X.; Zhang, Q.; Zheng, J.; Li, M.; Yu, M.; Wang, X.; Deng, M.; Zhai, X.; Li, R. Improved glucose and lipid metabolism in the early life of female offspring by maternal dietary genistein is associated with alterations in the gut microbiota. Front. Endocrinol. 2018, 9, 516. [Google Scholar] [CrossRef]
- Bui, T.P.N.; Shetty, S.A.; Lagkouvardos, I.; Ritari, J.; Chamlagain, B.; Douillard, F.P.; Paulin, L.; Piironen, V.; Clavel, T.; Plugge, C.M. Comparative genomics and physiology of the butyrate-producing bacterium Intestinimonas butyriciproducens. Environ. Microbiol. Rep. 2016, 8, 1024–1037. [Google Scholar] [CrossRef]
- Afouda, P.; Durand, G.A.; Lagier, J.C.; Labas, N.; Cadoret, F.; Armstrong, N.; Raoult, D.; Dubourg, G. Noncontiguous finished genome sequence and description of Intestinimonas massiliensis sp. nov strain GD 2T, the second Intestinimonas species cultured from the human gut. MicrobiologyOpen 2019, 8, e00621. [Google Scholar] [CrossRef] [PubMed]
- Afrizal, A.; Hitch, T.C.; Viehof, A.; Treichel, N.; Riedel, T.; Abt, B.; Buhl, E.M.; Kohlheyer, D.; Overmann, J.; Clavel, T. Anaerobic single-cell dispensing facilitates the cultivation of human gut bacteria. Environ. Microbiol. 2022, 24, 3861–3881. [Google Scholar] [CrossRef] [PubMed]
- Hamer, H.M.; Jonkers, D.; Venema, K.; Vanhoutvin, S.; Troost, F.; Brummer, R.J. The role of butyrate on colonic function. Aliment. Pharmacol. Ther. 2008, 27, 104–119. [Google Scholar] [CrossRef]
- Canfora, E.E.; Jocken, J.W.; Blaak, E.E. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat. Rev. Endocrinol. 2015, 11, 577–591. [Google Scholar] [CrossRef]
- Durand, G.; Cadoret, F.; Lagier, J.; Fournier, P.E.; Raoult, D. Description of ‘Gorbachella massiliensis’ gen. nov., sp. nov., ‘Fenollaria timonensis’ sp. nov., ‘Intestinimonas timonensis’ sp. nov. and ‘Collinsella ihuae’ sp. nov. isolated from healthy fresh stools with culturomics. New Microbes New Infect. 2017, 16, 60–62. [Google Scholar] [CrossRef]








| Phylum | Average | SD | Ratio | ava | avb | cumsum | p |
| Firmicutes | 0.028 | 0.020 | 1.435 | 62.132 | 57.000 | 0.365 | 0.001 |
| Bacteroidetes | 0.016 | 0.012 | 1.292 | 27.769 | 30.659 | 0.566 | 0.001 |
| Proteobacteria | 0.014 | 0.011 | 1.297 | 2.952 | 5.313 | 0.752 | 0.002 |
| Verrucomicrobia | 0.008 | 0.008 | 1.118 | 2.040 | 1.770 | 0.861 | 0.212 |
| Lentisphaerae | 0.003 | 0.003 | 0.990 | 2.346 | 2.872 | 0.904 | 0.004 |
| Spirochaetes | 0.003 | 0.002 | 1.231 | 1.355 | 0.871 | 0.939 | 0.001 |
| Actinobacteria | 0.002 | 0.003 | 0.637 | 0.268 | 0.448 | 0.965 | 0.173 |
| Kiritimatiellaeota | 0.001 | 0.001 | 1.375 | 0.511 | 0.524 | 0.977 | 0.429 |
| Fibrobacteres | 0.001 | 0.000 | 1.440 | 0.230 | 0.166 | 0.984 | 0.046 |
| Tenericutes | 0.000 | 0.000 | 1.478 | 0.150 | 0.167 | 0.988 | 0.173 |
| Genus | Average | SD | Ratio | ava | avb | cumsum | p |
| Clostridium | 0.020 | 0.024 | 0.835 | 4.053 | 1.138 | 0.091 | 0.081 |
| Streptococcus | 0.012 | 0.009 | 1.348 | 3.033 | 2.111 | 0.146 | 0.218 |
| Akkermansia | 0.008 | 0.008 | 1.083 | 1.975 | 1.616 | 0.184 | 0.24 |
| Escherichia | 0.008 | 0.012 | 0.658 | 0.432 | 1.622 | 0.220 | 0.011 |
| Rikenella | 0.007 | 0.004 | 1.634 | 5.053 | 6.400 | 0.252 | 0.001 |
| Succinivibrio | 0.006 | 0.009 | 0.742 | 0.006 | 1.276 | 0.281 | 0.001 |
| Lactobacillus | 0.006 | 0.010 | 0.577 | 1.549 | 1.411 | 0.308 | 0.227 |
| Ruminococcus | 0.006 | 0.003 | 2.233 | 2.078 | 0.926 | 0.334 | 0.001 |
| Intestinimonas | 0.005 | 0.003 | 1.400 | 8.486 | 9.076 | 0.355 | 0.028 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, P.; Gao, W.; Cong, S.; Leng, L.; Wang, T.; Shi, L. High-Energy Supplemental Feeding Shifts Gut Microbiota Composition and Function in Red Deer (Cervus elaphus). Animals 2024, 14, 1428. https://doi.org/10.3390/ani14101428
Zheng P, Gao W, Cong S, Leng L, Wang T, Shi L. High-Energy Supplemental Feeding Shifts Gut Microbiota Composition and Function in Red Deer (Cervus elaphus). Animals. 2024; 14(10):1428. https://doi.org/10.3390/ani14101428
Chicago/Turabian StyleZheng, Peng, Weizhen Gao, Shaobo Cong, Lin Leng, Tao Wang, and Lei Shi. 2024. "High-Energy Supplemental Feeding Shifts Gut Microbiota Composition and Function in Red Deer (Cervus elaphus)" Animals 14, no. 10: 1428. https://doi.org/10.3390/ani14101428





