Physiological and Transcriptome Analyses Reveal the Protective Effect of Exogenous Trehalose in Response to Heat Stress in Tea Plant (Camellia sinensis)
Abstract
:1. Introduction
2. Results
2.1. Effects of Exogenous Trehalose on Growth Morphology of Tea Plant under Heat Stress
2.2. Effects of Exogenous Trehalose on Physiological Characteristics of Tea Plant Response to Heat Stress
2.3. Transcriptome Analysis and Differentially Expressed Genes Identification for Heat Stress Response after Exogenous Trehalose Treatment in Tea Plant
2.4. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Pathway Analyses of DEGs in C. sinensis under Heat Stress Regulated by Exogenous Trehalose
2.5. Analysis of DEGs Involved in MAPK Signaling and Plant Hormone Signal Transduction Pathways in Tea Plants under Heat Stress with Exogenous Trehalose Treatment
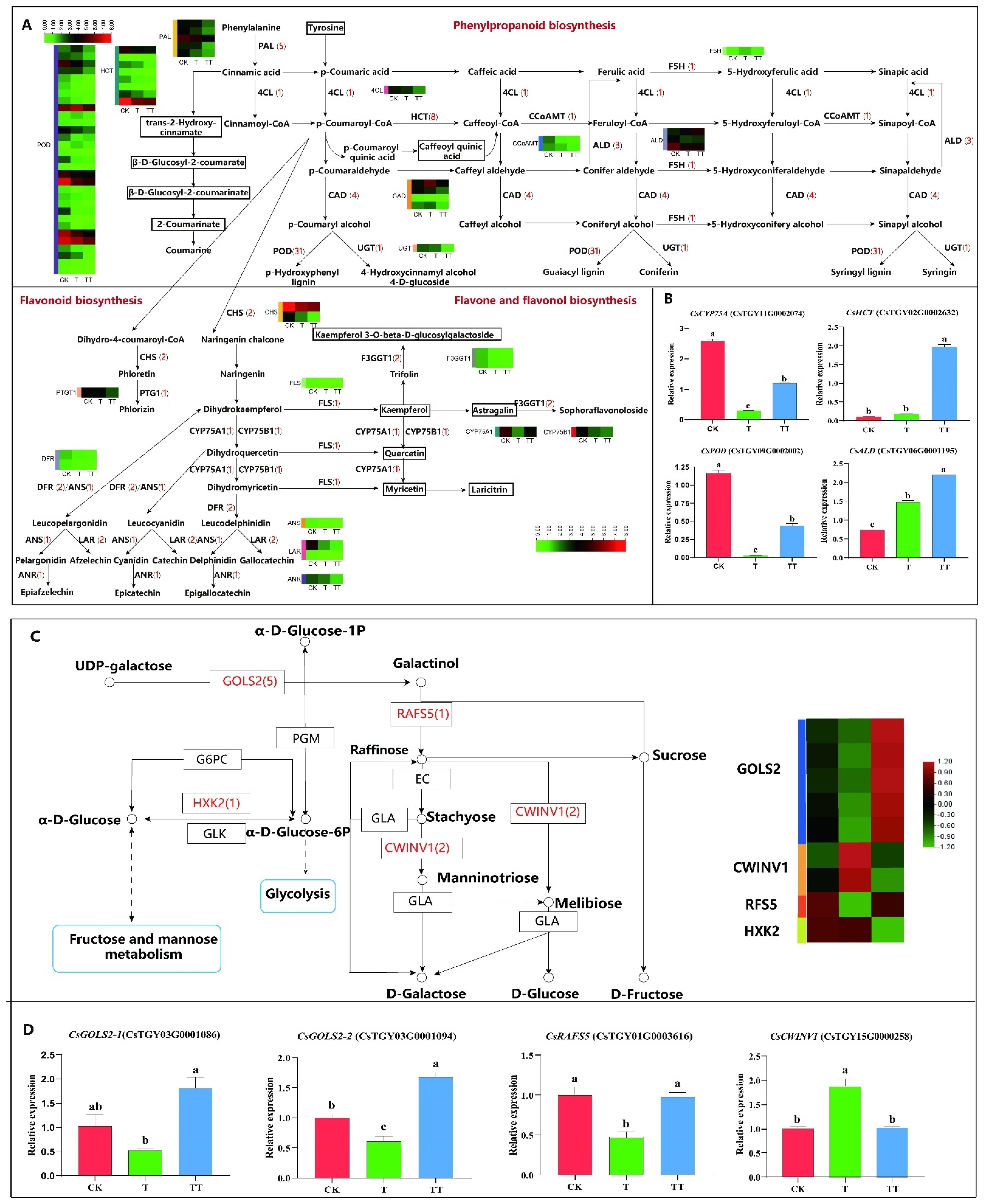
2.6. Analysis of DEGs Involved in Phenylpropanoid Biosynthesis, Flavonoid Biosynthesis, and Flavone and Flavonol Biosynthesis in Tea Plants under Heat Stress with Exogenous Trehalose Treatment
2.7. Analysis of DEGs Involved in Galactose Metabolism in Tea Plants under Heat Stress with Exogenous Trehalose Treatment
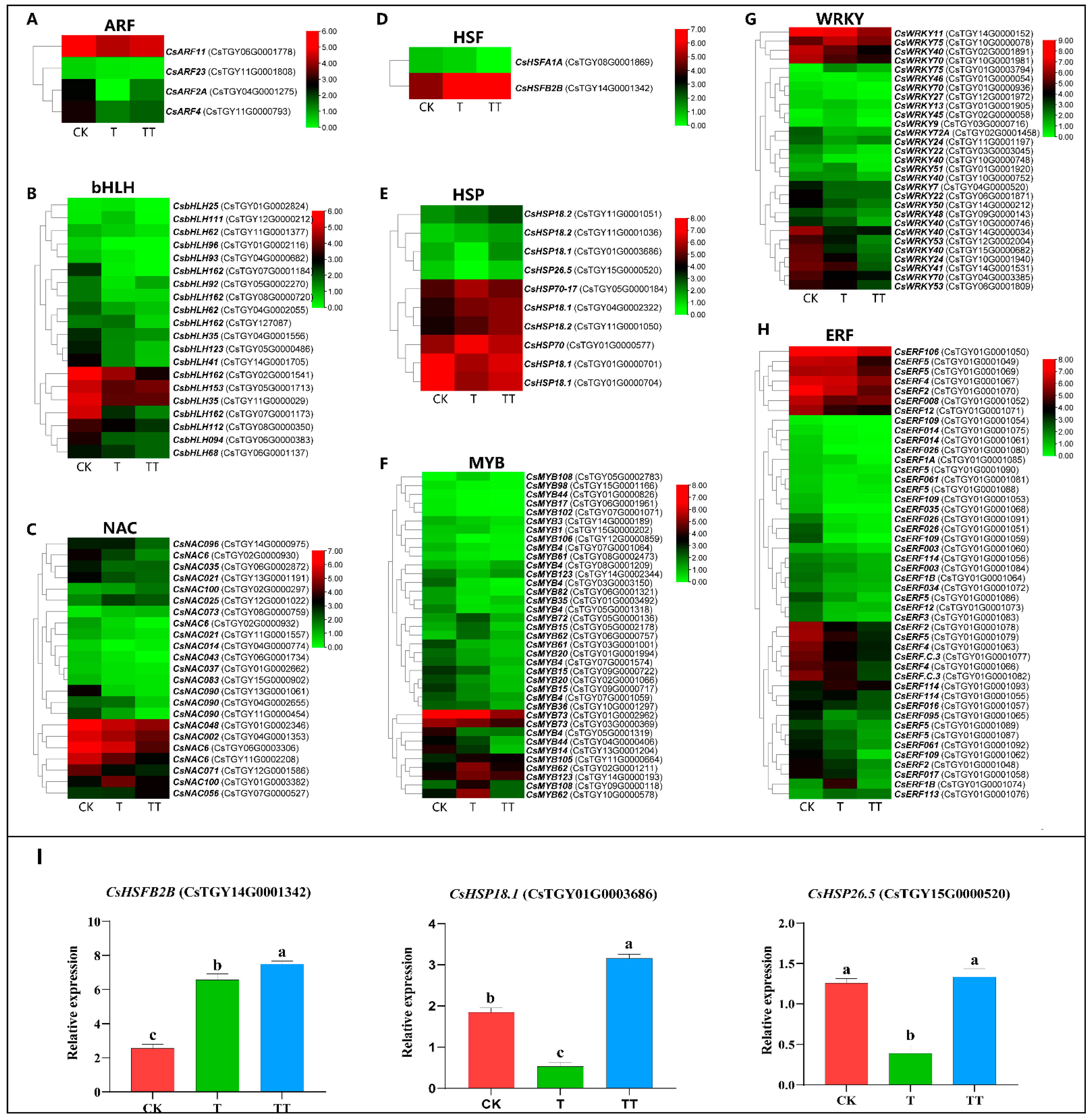
2.8. Analysis of Transcription Factors in Response to Heat Stress under Exogenous Trehalose Treatment in Tea Plants
3. Discussion
3.1. Trehalose Alleviates the Damage Caused by Heat Stress through Increasing the Activity of Antioxidant Enzymes and the Content of Osmotic Substance
3.2. Trehalose May Enhance the Heat Tolerance of Tea Plants by Activating the ABA and MAPK Signal Transduction Pathway
3.3. Trehalose Could Induce Gene Expression in the Galactose Pathway of Tea Plants under Heat Stress and Improve the Heat Resistance of Tea Plants
3.4. Trehalose Enhances the Heat Resistance of Tea Plants by Inducing the Expression of Key Genes Involved in Lignin Synthesis in the Phenylpropanoid Pathway
3.5. Trehalose Improves the Heat Tolerance of the Tea Plant by Inducing the Expression of HSF and HSP under Heat Stress
4. Materials and Methods
4.1. Plant Materials and Heat Stress Treatment
4.2. Determination of Antioxidant Enzyme Activity
4.3. Determination of MDA, Proline, Soluble Sugar, and Trehalose Contents
4.4. RNA Extraction and Transcriptome Sequencing
4.5. RNA-Seq Data Analysis
4.6. Different Expression Analysis
4.7. Quantitative Real-Time PCR (qRT-PCR) Analysis
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Xia, E.H.; Tong, W.; Wu, Q.; Wei, S.; Zhao, J.; Zhang, Z.Z.; Wei, C.L.; Wan, X.C. Tea plant genomics: Achievements, challenges and perspectives. Hortic. Res. 2020, 7, 2746–2764. [Google Scholar] [CrossRef]
- Li, X.; Wei, J.P.; Ahammed, G.J.; Zhang, L.; Li, Y.; Yan, P.; Zhang, L.P.; Han, W.Y. Brassinosteroids Attenuate Moderate High Temperature-Caused Decline in Tea Quality by Enhancing Theanine Biosynthesis in Camellia sinensis L. Front. Plant Sci. 2018, 9, 1016. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Song, Y.; Li, H.; Zaman, S.; Fan, K.; Ding, Z.; Wang, Y. Enhancing the Adaptability of Tea Plants (Camellia sinensis L.) to High-Temperature Stress with Small Peptides and Biosurfactants. Plants 2023, 12, 2817. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Wang, Y.; Qiu, C.; Qian, W.; Xie, H.; Ding, Z. Alternative splicing in tea plants was extensively triggered by drought, heat and their combined stresses. PeerJ 2020, 8, e8258. [Google Scholar] [CrossRef]
- Welch, J.R.; Vincent, J.R.; Auffhammer, M.; Moya, P.F.; Dobermann, A.; Dawe, D. Rice yields in tropical/subtropical Asia exhibit large but opposing sensitivities to minimum and maximum temperatures. Proc. Natl. Acad. Sci. USA 2010, 107, 14562–14567. [Google Scholar] [CrossRef]
- Kumar, S.; Thakur, M.; Mitra, R.; Basu, S.; Anand, A. Sugar metabolism during pre- and post-fertilization events in plants under high temperature stress. Plant Cell Rep. 2022, 41, 55–673. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, D.; Zhou, L.; Zhang, X.; Liao, J.; Duan, Y.; Wen, B.; Ma, Y.; Wang, Y.; Fang, W.; et al. Transcriptomic and metabolomic profiling of Camellia sinensis L. cv. ‘Suchazao’ exposed to temperature stresses reveals modification in protein synthesis and photosynthetic and anthocyanin biosynthetic pathways. Tree Physiol. 2019, 39, 1583–1599. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, S.; Dard, A.; Meyer, A.J.; Reichheld, J.P. Redox-mediated responses to high temperature in plants. J. Exp. Bot. 2023, 74, 2489–2507. [Google Scholar] [CrossRef]
- Jong, K.S.; Hui, Y.L.; Yu, C.M.; Ki, S.Y.; Kim, S.H.; Pak, H.H. The impact of cryoprotective media on cryopreservation of cells using loading trehalose. Cryobiology 2020, 92, 258–259. [Google Scholar] [CrossRef]
- Zhu, F.; Li, M.; Sun, M.; Jiang, X.; Qiao, F. Plant hormone signals regulate trehalose accumulation against osmotic stress in watermelon cells. Protoplasma 2022, 259, 1351–1369. [Google Scholar] [CrossRef]
- Sharma, E.; Shruti, P.S.; Singh, S.; Singh, T.; Kaur, P.; Jodha, B.; Srivastava, Y.; Munshi, A.; Singh, S. Trehalose and its Diverse Biological Potential. Curr. Protein Pept. Sci. 2023, 24, 503–517. [Google Scholar] [CrossRef]
- Redillas, M.C.F.R.; Park, S.H.; Lee, J.W.; Kim, Y.S.; Jeong, J.S.; Jung, H.; Bang, S.W.; Hahn, T.R.; Kim, J.K. Accumulation of trehalose increases soluble sugar contents in rice plants conferring tolerance to drought and salt stress. Plant Biotechnol. Rep. 2012, 6, 89–96. [Google Scholar] [CrossRef]
- Sarkar, A.K.; Sadhukhan, S. Imperative role of trehalose metabolism and trehalose-6-phosphate signaling on salt stress responses in plants. Physiol. Plant. 2022, 174, e13647. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, H.L.; Du, J.; Du, G.; Zhan, J.C.; Huang, W.D. Trehalose protects wine yeast against oxidation under thermal stress. World J. Microb. Biot. 2010, 26, 969–976. [Google Scholar] [CrossRef]
- Zhang, G.; Yan, P.; Leng, D.; Shang, L.; Zhang, C.; Wu, Z.; Wang, Z. Salicylic Acid Treatment Alleviates the Heat Stress Response by Reducing the Intracellular ROS Level and Increasing the Cytosolic Trehalose Content in Pleurotus ostreatus. Microbiol. Spectr. 2023, 11, e03113–e03122. [Google Scholar] [CrossRef] [PubMed]
- Shirasaka, N.; Yoshida, K.; Terashima, Y.; Watanabe, T.; Ayusawa, S.; Terashita, T. Effect of trehalose on the heat tolerance of Lentinula edodes mycelia. Mushroom Sci. Biotechnol. 2006, 14, 11–18. [Google Scholar]
- Zhang, Z.; Sun, M.; Gao, Y.; Luo, Y. Exogenous trehalose differently improves photosynthetic carbon assimilation capacities in maize and wheat under heat stress. J. Plant Interact. 2022, 17, 361–370. [Google Scholar] [CrossRef]
- Luo, Y.; Liu, X.Y.; Fan, Y.Z.; Fan, Y.H.; Lv, Z.Y.; Li, W.Q.; Cen, J.Y. Exogenously Supplied Trehalose Accelerates Photosynthetic Electron Transport in Heat-Stressed Maize. Russ. J. Plant Physiol. 2021, 68, 857–866. [Google Scholar] [CrossRef]
- Luo, Y.; Wang, W.; Fan, Y.Z.; Gao, Y.M.; Wang, D. Exogenously-Supplied Trehalose Provides Better Protection for D1 Protein in Winter Wheat under Heat Stress. Russ. J. Plant Physiol. 2018, 65, 115–122. [Google Scholar] [CrossRef]
- Luo, Y.; Liu, H.Y.; Fan, Y.Z.; Wang, W.; Zhao, Y.Y. Comparative chloroplast proteome analysis of exogenously supplied trehalose to wheat seedlings under heat stress. Photosynthetica 2018, 56, 1123–1133. [Google Scholar] [CrossRef]
- Zhao, D.Q.; Li, T.T.; Hao, Z.J.; Cheng, M.L.; Tao, J. Exogenous trehalose confers high temperature stress tolerance to herbaceous peony by enhancing antioxidant systems, activating photosynthesis, and protecting cell structure. Cell Stress Chaperon. 2019, 24, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, S.K.; Basu, S.; Kumar, S.; Kumari, S.; Kumar, A.; Jha, S.; Mishra, J.S.; Bhatt, B.P.; Kumar, G. Enhanced antioxidant enzyme activities in developing anther contributes to heat stress alleviation and sustains grain yield in wheat. Funct. Plant Biol. 2019, 46, 1090–1102. [Google Scholar] [CrossRef] [PubMed]
- Uzal, O.; Baslak, L.; Yasar, F. Effects of External Melatonin Treatments on Morphological and Physiological Changes in Cucumber (Cucumis sativus L.) Seedlings Against Chilling Stress. Gesunde Pflanz. 2023, 75, 115–125. [Google Scholar] [CrossRef]
- Li, R.; Xie, Y.; Tang, Y. Effect of Application Accumulator Plant Straw on the Osmotic Adjustment Substances and Malondialdehyde Content of Lettuce. IOP Conf. Ser. Earth Environ. Sci. 2019, 233, 042025. [Google Scholar] [CrossRef]
- Kavi Kishor, P.B.; Suravajhala, P.; Rathnagiri, P.; Sreenivasulu, N. Intriguing Role of Proline in Redox Potential Conferring High Temperature Stress Tolerance. Front. Plant Sci. 2022, 13, 867531. [Google Scholar] [CrossRef]
- Luo, Y.; Wang, Y.; Xie, Y.; Gao, Y.; Li, W.; Lang, S. Transcriptomic and Metabolomic Analyses of the Effects of Exogenous Trehalose on Heat Tolerance in Wheat. Int. J. Mol. Sci. 2022, 23, 5194. [Google Scholar] [CrossRef] [PubMed]
- Nowicka, B. Modifications of Phytohormone Metabolism Aimed at Stimulation of Plant Growth, Improving Their Productivity and Tolerance to Abiotic and Biotic Stress Factors. Plants 2022, 11, 3430. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Zandalinas, S.I.; Balfagón, D.; Arbona, V.; Gómez-Cadenas, A.; Inupakutika, M.A.; Mittler, R. ABA is required for the accumulation of APX1 and MBF1c during a combination of water deficit and heat stress. J. Exp. Bot. 2016, 67, 5381–5390. [Google Scholar] [CrossRef]
- Lata, C.; Prasad, M. Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 2011, 62, 4731–4748. [Google Scholar] [CrossRef]
- Xu, P.; Zhang, X.; Su, H.; Liu, X.; Wang, Y.; Hong, G. Genome-wide Analysis of PYL-PP2C-SnRK2s Family in Camellia sinensis. Bioengineered 2020, 11, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, L.; Sheng, X.; Yan, C.; Zhou, R.; Hang, J.; Yin, P.; Yan, N. Molecular basis for the selective and ABA-independent inhibition of PP2CA by PYL13. Cell Res. 2013, 23, 1369–1379. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, S.; Shi, L.; Gong, D.; Zhang, S.; Zhao, Q.; Zhan, D.; Vasseur, L.; Wang, Y.; Yu, J.; et al. Haplotype-resolved genome assembly provides insights into evolutionary history of the tea plant Camellia sinensis. Nat. Genet. 2021, 53, 1250–1259. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, Y.; Duan, C.; Chen, P.; Li, Q.; Dai, S.; Sun, L.; Ji, K.; Sun, Y.; Xu, W.; et al. The expression profiling of the CsPYL, CsPP2C and CsSnRK2 gene families during fruit development and drought stress in cucumber. J. Plant Physiol. 2012, 169, 1874–1882. [Google Scholar] [CrossRef] [PubMed]
- Dalal, M.; Inupakutika, M. Transcriptional regulation of ABA core signaling component genes in sorghum (Sorghum bicolor L. Moench). Mol. Breed. 2014, 34, 1517–1525. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Baena-González, E.; Sheen, J. Convergent energy and stress signaling. Trends Plant Sci. 2008, 13, 474–482. [Google Scholar] [CrossRef] [PubMed]
- Jing, Y.; Lang, S.; Wang, D.; Xue, H.; Wang, X.F. Functional characterization of galactinol synthase and raffinose synthase in desiccation tolerance acquisition in developing Arabidopsis seeds. J. Plant Physiol. 2018, 230, 109–121. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, T.B.; Budzinski, I.G.; Marur, C.J.; Petkowicz, C.L.; Pereira, L.F.; Vieira, L.G. Expression of three galactinol synthase isoforms in Coffea arabica L. and accumulation of raffinose and stachyose in response to abiotic stresses. Plant Physiol. Biochem. 2011, 49, 441–448. [Google Scholar] [CrossRef]
- Cui, R.; Wang, X.; Malik, W.A.; Lu, X.; Chen, X.; Wang, D.; Wang, J.; Wang, S.; Chen, C.; Guo, L.; et al. Genome-wide identification and expression analysis of Raffinose synthetase family in cotton. BMC Bioinform. 2021, 22, 356–368. [Google Scholar] [CrossRef]
- Yang, J.; Ling, C.; Liu, Y.; Zhang, H.; Hussain, Q.; Lyu, S.; Wang, S.; Liu, Y. Genome-Wide Expression Profiling Analysis of Kiwifruit GolS and RFS Genes and Identification of AcRFS4 Function in Raffinose Accumulation. Int. J. Mol. Sci. 2022, 23, 8836. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, A.; Yabuta, Y.; Shigeoka, S. Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol. 2008, 147, 1251–1263. [Google Scholar] [CrossRef] [PubMed]
- Lang, S.; Liu, X.; Xue, H.; Li, X.; Wang, X. Functional characterization of BnHSFA4a as a heat shock transcription factor in controlling the re-establishment of desiccation tolerance in seeds. J. Exp. Bot. 2017, 68, 2361–2375. [Google Scholar] [CrossRef] [PubMed]
- Vanholme, R.; De Meester, B.; Ralph, J.; Boerjan, W. Lignin biosynthesis and its integration into metabolism. Curr. Opin. Biotechnol. 2019, 56, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and Biological Functions in Plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef] [PubMed]
- Rao, X.; Barros, J. Modeling lignin biosynthesis: A pathway to renewable chemicals. Trends Plant Sci. 2023, 29, S1360–S1385. [Google Scholar] [CrossRef]
- Wang, Y.X.; Teng, R.M.; Wang, W.L.; Wang, Y.; Shen, W.; Zhuang, J. Identification of genes revealed differential expression profiles and lignin accumulation during leaf and stem development in tea plant (Camellia sinensis (L.) O. Kuntze). Protoplasma 2019, 256, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.; Wang, L.; Zhang, C.; Wu, L.; Li, H.; Zhang, F.; Cheng, H. Transcriptome Analysis Reveals Key Flavonoid 3′-Hydroxylase and Flavonoid 3′,5′-Hydroxylase Genes in Affecting the Ratio of Dihydroxylated to Trihydroxylated Catechins in Camellia sinensis. PLoS ONE 2015, 10, e0137925. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhao, Y.; Wang, L.; Hou, Y.; Bao, Y.; Jia, Z.; Zheng, Y.; Jin, P. Hot water treatment improves peach fruit cold resistance through PpHSFA4c-mediated HSF-HSP and ROS pathways. Postharvest Biol. Technol. 2023, 199, 112272. [Google Scholar] [CrossRef]
- Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol. 2011, 157, 1243–1254. [Google Scholar] [CrossRef]
- Kolmos, E.; Chow, B.Y.; Pruneda-Paz, J.L.; Kay, S.A. HsfB2b-mediated repression of PRR7 directs abiotic stress responses of the circadian clock. Proc. Natl. Acad. Sci. USA 2014, 111, 16172–16177. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zou, Z.; Li, Q.; Xin, H.; Zhu, X.; Chen, X.; Li, X. Heterologous expression of three Camellia sinensis small heat shock protein genes confers temperature stress tolerance in yeast and Arabidopsis thaliana. Plant Cell Rep. 2017, 36, 1125–1135. [Google Scholar] [CrossRef] [PubMed]
- Vianna, C.R.; Ferreira, M.C.; Silva, C.L.; Tanghe, A.; Neves, M.J.; Thevelein, J.M.; Rosa, C.A.; Van Dijck, P. Stress tolerance of the Saccharomyces cerevisiae adenylate cyclase fil1 (CYR1lys1682) mutant depends on Hsp26. J. Mol. Microbiol. Biotechnol. 2010, 19, 140–146. [Google Scholar] [PubMed]
- Roberts, G.R.; Keys, A.J. The mechanism of photosynthesis in the tea plant (Camellia sinensis L.). J. Exp. Bot. 1978, 29, 1403–1407. [Google Scholar] [CrossRef]
- Barman, T.S.; Baruah, U.; Saikia, J.K. Irradiance influences tea leaf (Camellia sinensis L.) photosynthesis and transpiration. Photosynthetica 2008, 46, 618–621. [Google Scholar] [CrossRef]
- Seth, R.; Maritim, T.K.; Parmar, R.; Sharma, R.K. Underpinning the molecular programming attributing heat stress associated thermotolerance in tea (Camellia sinensis (L.) O. Kuntze). Hortic. Res. 2021, 8, 99–117. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Li, Z.; Yang, J.; Zu, Y. Responses of antioxidant enzymes and key resistant substances in perennial ryegrass (Lolium perenne L.) to cadmium and arsenic stresses. BMC Plant Biol. 2022, 22, 145–159. [Google Scholar] [CrossRef] [PubMed]
- Heath, R.L.; Packer, L. Reprint of: Photoperoxidation in isolated chloroplasts: I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 2022, 726, 109248. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Yi, X.; Gao, X.; Wang, M.; Shao, C.; Lv, Z.; Chen, J.; Liu, Z.; Shen, C. Physiological and biochemical responses of tea seedlings (Camellia sinensis) to simulated acid rain conditions. Ecotoxicol. Environ. Saf. 2020, 192, 110315. [Google Scholar] [CrossRef]
- Li, Y.; Lv, Y.; Lian, M.; Peng, F.; Xiao, Y. Effects of combined glycine and urea fertilizer application on the photosynthesis, sucrose metabolism, and fruit development of peach. Sci. Hortic. 2021, 289, 110504. [Google Scholar] [CrossRef]
- Huang, Q.; Zhang, G.; Nan, J.; Cheng, W.; Zhu-Salzman, K. Characterization of trehalose metabolic genes and corresponding enzymatic activities during diapause of Sitodiplosis mosellana. J. Insect Physiol. 2021, 135, 104324. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, S.; Li, W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics 2012, 28, 2184–2185. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.; Pimentel, H.; Trapnell, C.; Pachter, L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 2011, 27, 2325–2329. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential Expression of RNA-Seq Data at the Gene Level–The DESeq Package; European Molecular Biology Laboratory (EMBL): Heidelberg, Germany, 2012; pp. 1–23. [Google Scholar]
- Gene Ontology Consortium. The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006, 34, D322–D326. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Arocho, A.; Chen, B.; Ladanyi, M.; Pan, Q. Validation of the 2−ΔΔCt Calculation as an Alternate Method of Data Analysis for Quantitative PCR of BCR-ABL P210 Transcripts. Diagn. Mol. Pathol. 2006, 15, 56–61. [Google Scholar] [CrossRef]




| Sample | Total Raw Reads (Mb) | Clean Reads (Mb) | Clean Bases (Gb) | Q30(%) | GC (%) | Total Mapped Reads (%) |
|---|---|---|---|---|---|---|
| CK-1 | 117.79 | 113.10 | 16.96 | 91.52% | 44.02% | 74.45 |
| CK-2 | 134.47 | 130.16 | 19.52 | 91.50% | 45.71% | 76.28 |
| CK-3 | 126.45 | 121.31 | 18.20 | 91.51% | 43.92% | 74.58 |
| T-1 | 117.20 | 113.68 | 17.05 | 91.54% | 45.52% | 80.95 |
| T-2 | 119.72 | 115.70 | 17.36 | 91.65% | 44.37% | 80.28 |
| T-3 | 107.14 | 103.24 | 15.49 | 91.71% | 44.04% | 80.17 |
| TT-1 | 125.50 | 119.94 | 17.99 | 91.54% | 44.34% | 74.09 |
| TT-2 | 112.49 | 107.32 | 16.10 | 91.56% | 44.14% | 73.65 |
| TT-3 | 111.43 | 106.45 | 15.97 | 92.02% | 44.28% | 73.59 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, S.; Liu, C.; Zhou, Z.; Xu, L.; Lai, Z. Physiological and Transcriptome Analyses Reveal the Protective Effect of Exogenous Trehalose in Response to Heat Stress in Tea Plant (Camellia sinensis). Plants 2024, 13, 1339. https://doi.org/10.3390/plants13101339
Zheng S, Liu C, Zhou Z, Xu L, Lai Z. Physiological and Transcriptome Analyses Reveal the Protective Effect of Exogenous Trehalose in Response to Heat Stress in Tea Plant (Camellia sinensis). Plants. 2024; 13(10):1339. https://doi.org/10.3390/plants13101339
Chicago/Turabian StyleZheng, Shizhong, Chufei Liu, Ziwei Zhou, Liyi Xu, and Zhongxiong Lai. 2024. "Physiological and Transcriptome Analyses Reveal the Protective Effect of Exogenous Trehalose in Response to Heat Stress in Tea Plant (Camellia sinensis)" Plants 13, no. 10: 1339. https://doi.org/10.3390/plants13101339





