The Expression of Adipogenic Marker Is Significantly Increased in Estrogen-Treated Lipedema Adipocytes Differentiated from Adipose Stem Cells In Vitro
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Treatment of ASCs with 17β-Estradiol
2.3. AlamarBlue Cell Proliferation Assay
2.4. Flow Cytometry
2.5. Adipogenic Differentiation
2.6. Formation of ASC Spheroids
2.7. Quantitative Polymerase Chain Reaction (qPCR)
2.8. Western Blot Analyses
2.9. Statistical Analysis
3. Results
3.1. Estrogen Increased the Proliferation and the Expression of Stemness Markers in Healthy ASCs in 2D Monolayer Culture
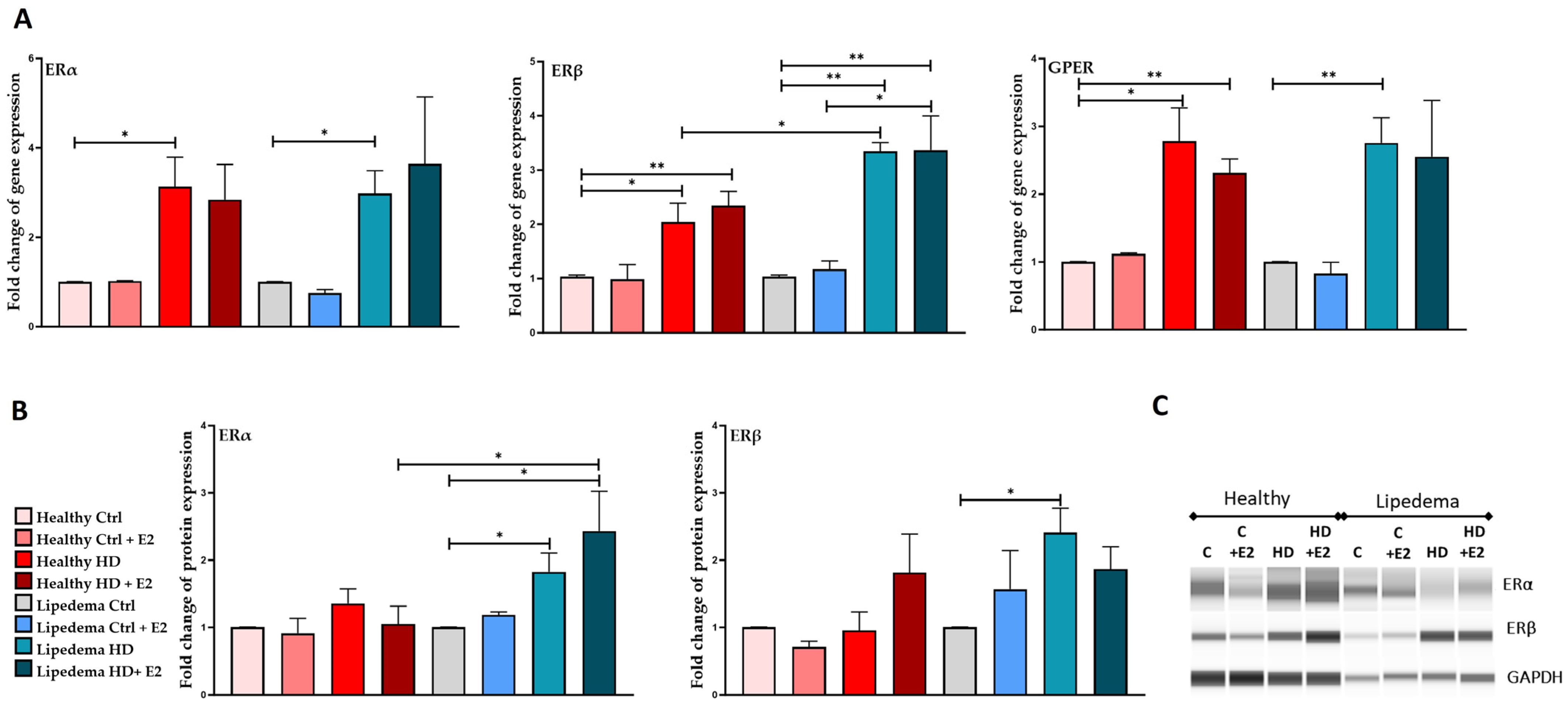
3.2. Estrogen Increased the Expression of ERs in ASCs in 2D Monolayer Culture, but It Decreased the Expression of ERα and GPER in Lipedema Spheroid
3.3. Estrogen Significantly Increased the Expression of HSD17B7, LIPE, and STS in Lipedema ASCs and CYP19A1 in Healthy ASCs in 2D Monolayer Culture
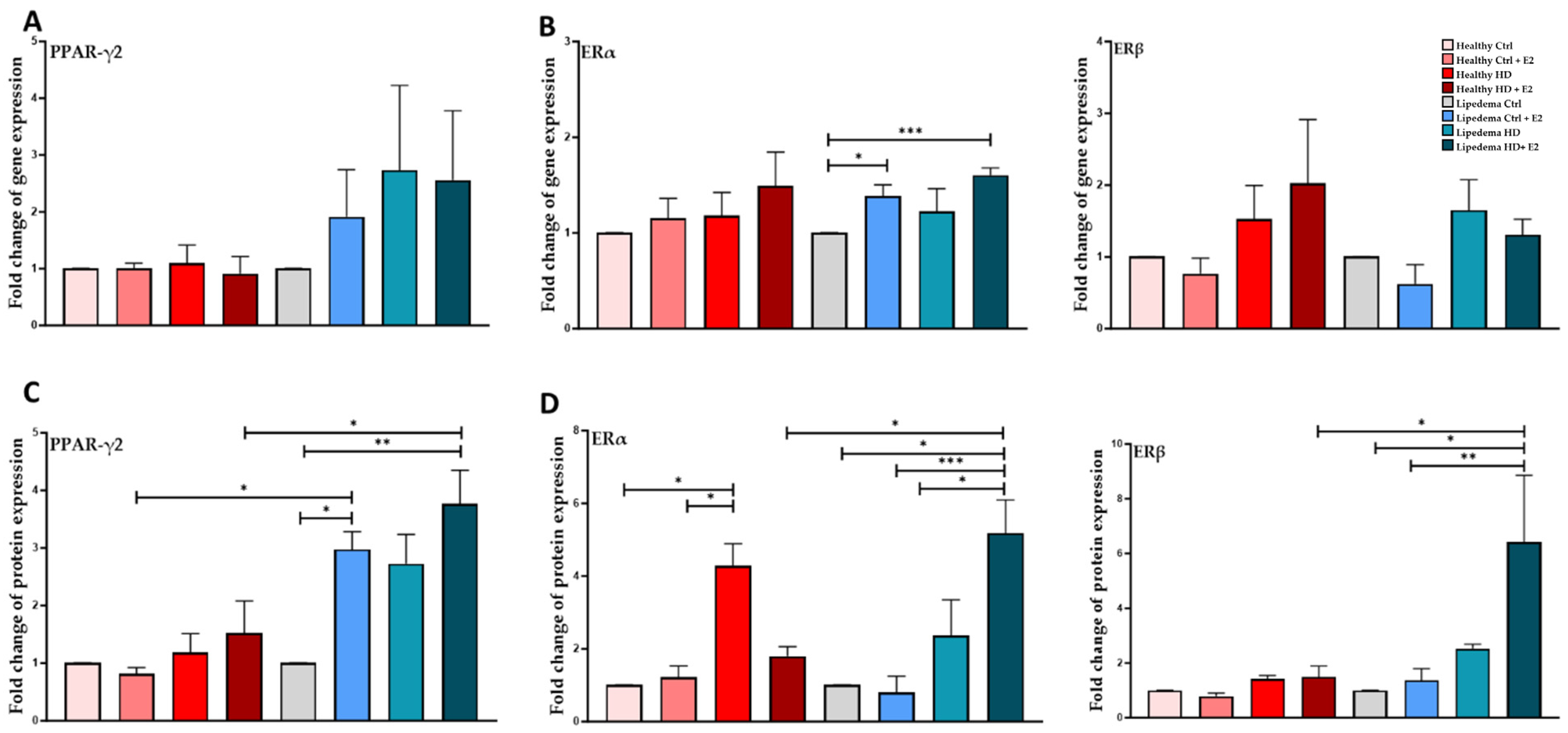
3.4. Estrogen Significantly Increased the Expression of PPAR-ϒ2 in Differentiated Lipedema Adipocytes and Spheroids
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Herbst, K.L.; Kahn, L.A.; Iker, E.; Ehrlich, C.; Wright, T.; McHutchison, L.; Schwartz, J.; Sleigh, M.; MC Donahue, P.; Lisson, K.H.; et al. Standard of care for lipedema in the United States. Phlebology 2021, 36, 779–796. [Google Scholar] [CrossRef] [PubMed]
- Wold, L.E.; Hines, E.A., Jr.; Allen, E.V. Lipedema of the legs is a syndrome characterized by fat legs and edema. Ann. Intern. Med. 1951, 34, 1243–1250. [Google Scholar] [PubMed]
- Herbst, K.L. Subcutaneous Adipose Tissue Diseases: Dercum, Lipedema, Familial Multiple Lipomatosis, and Madelung Disease. In Endotext; Feingold, K.R., Anawalt, B., Blackman, M.R., Boyce, A., Chrousos, G., Corpas, E., de Herder, W.W., Dhatariya, K., Dungan, K., Hofland, J., et al., Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar]
- Poojari, A.; Dev, K.; Rabiee, A. Lipedema: Insights into Morphology, Pathophysiology, and Challenges. Biomedicines 2022, 10, 3081. [Google Scholar] [CrossRef]
- Buck, D.W., 2nd; Herbst, K.L. Lipedema: A Relatively Common Disease with Extremely Common Misconceptions. Plast. Reconstr. Surg. Glob. Open 2016, 4, e1043. [Google Scholar] [CrossRef]
- Schmeller, W.; Hueppe, M.; Meier-Vollrath, I. Tumescent liposuction in lipoedema yields good long-term results. Br. J. Dermatol. 2012, 166, 161–168. [Google Scholar] [CrossRef]
- Kirstein, F.; Hamatschek, M.; Knors, H.; Aitzetmueller-Klietz, M.-L.; Aitzetmueller-Klietz, M.; Wiebringhaus, P.; Varnava, C.; Hirsch, T.; Kueckelhaus, M. Patient-Reported Outcomes of Liposuction for Lipedema Treatment. Healthcare 2023, 11, 2020. [Google Scholar] [CrossRef]
- Dadras, M.; Mallinger, P.J.; Corterier, C.C.; Theodosiadi, S.; Ghods, M. Liposuction in the Treatment of Lipedema: A Longitudinal Study. Arch. Plast. Surg. 2017, 44, 324–331. [Google Scholar] [CrossRef]
- Al-Ghadban, S.; Walczak, S.G.; Isern, S.U.; Martin, E.C.; Herbst, K.L.; Bunnell, B.A. Enhanced Angiogenesis in HUVECs Preconditioned with Media from Adipocytes Differentiated from Lipedema Adipose Stem Cells In Vitro. Int. J. Mol. Sci. 2023, 24, 13572. [Google Scholar] [CrossRef] [PubMed]
- Al-Ghadban, S.; Cromer, W.; Allen, M.; Ussery, C.; Badowski, M.; Harris, D.; Herbst, K.L. Dilated Blood and Lymphatic Microvessels, Angiogenesis, Increased Macrophages, and Adipocyte Hypertrophy in Lipedema Thigh Skin and Fat Tissue. J. Obes. 2019, 2019, 8747461. [Google Scholar] [CrossRef]
- Suga, H.; Araki, J.; Aoi, N.; Kato, H.; Higashino, T.; Yoshimura, K. Adipose tissue remodeling in lipedema: Adipocyte death and concurrent regeneration. J. Cutan. Pathol. 2009, 36, 1293–1298. [Google Scholar] [CrossRef]
- Felmerer, G.; Stylianaki, A.; Hägerling, R.; Wang, A.; Ströbel, P.; Hollmén, M.; Lindenblatt, N.; Gousopoulos, E. Adipose Tissue Hypertrophy, An Aberrant Bio-chemical Profile and Distinct Gene Expression in Lipedema. J. Surg. Res. 2020, 253, 294–303. [Google Scholar] [CrossRef] [PubMed]
- Katzer, K.; Hill, J.L.; McIver, K.B.; Foster, M.T. Lipedema and the Potential Role of Estrogen in Excessive Adipose Tissue Accumulation. Int. J. Mol. Sci. 2021, 22, 11720. [Google Scholar] [CrossRef] [PubMed]
- Kaftalli, J.; Bonetti, G.; Marceddu, G.; Dhuli, K.; E Maltese, P.; Donato, K.; Herbst, K.L.; Michelini, S.; Chiurazzi, P.; Hill, M.; et al. AKR1C1 and hormone metabolism in lipedema pathogenesis: A computational biology approach. Eur. Rev. Med. Pharmacol. Sci. 2023, 27 (Suppl. S6), 137–147. [Google Scholar] [PubMed]
- Gavin, K.M.; Cooper, E.E.; Hickner, R.C. Estrogen receptor protein content is different in the abdominal than in gluteal subcutaneous adipose tissue of overweight-to-obese. Metabolism 2013, 62, 1180–1188. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Prossnitz, E.R. G-Protein-Coupled Estrogen Receptor (GPER) and Sex-Specific Metabolic Homeostasis. Adv. Exp. Med. Biol. 2017, 1043, 427–453. [Google Scholar] [PubMed]
- Lapid, K.; Lim, A.; Clegg, D.J.; Zeve, D.; Graff, J.M. Oestrogen signalling in white adipose progenitor cells inhibits differentiation into brown adipose and smooth muscle cells. Nat. Commun. 2014, 5, 5196. [Google Scholar] [CrossRef] [PubMed]
- Foryst-Ludwig, A.; Clemenz, M.; Hohmann, S.; Hartge, M.; Sprang, C.; Frost, N.; Krikov, M.; Bhanot, S.; Barros, R.; Morani, A.; et al. Metabolic actions of estrogen receptor beta (ERbeta) are mediated by a negative cross-talk with PPARgamma. PLoS Genet. 2008, 4, e1000108. [Google Scholar] [CrossRef]
- Davis, K.E.; Neinast, M.D.; Sun, K.; Skiles, W.M.; Bills, J.D.; Zehr, J.A.; Zeve, D.; Hahner, L.D.; Cox, D.W.; Gent, L.M.; et al. The sexually dimorphic role of adipose and adipocyte estrogen receptors in modulating adipose tissue expansion, inflammation, and fibrosis. Mol. Metab. 2013, 2, 227–242. [Google Scholar] [CrossRef] [PubMed]
- Mauvais-Jarvis, F.; Clegg, D.J.; Hevener, A.L. The Role of Estrogens in Control of Energy Balance and Glucose Homeostasis. Endocr. Rev. 2013, 34, 309–338. [Google Scholar] [CrossRef]
- Pallottini, V.; Bulzomi, P.; Galluzzo, P.; Martini, C.; Marino, M. Estrogen Regulation of Adipose Tissue Functions: Involvement of Estrogen Receptor Isoforms. Infect. Disord.-Drug Targets 2008, 8, 52–60. [Google Scholar] [CrossRef]
- Fatima, L.A.; Campello, R.S.; Santos, R.d.S.; Freitas, H.S.; Frank, A.P.; Machado, U.F.; Clegg, D.J. Estrogen receptor 1 (ESR1) regulates VEGFA in adipose tissue. Sci. Rep. 2017, 7, 16716. [Google Scholar] [CrossRef]
- Felmerer, G.; Stylianaki, A.; Hollmén, M.; Ströbel, P.; Stepniewski, A.; Wang, A.; Frueh, F.S.; Kim, B.-S.; Giovanoli, P.; Lindenblatt, N.; et al. Increased levels of VEGF-C and macrophage infiltration in lipedema patients without changes in lymphatic vascular morphology. Sci. Rep. 2020, 10, 10947. [Google Scholar] [CrossRef]
- Jouffre, B.; Acramel, A.; Jacquot, Y.; Daulhac, L.; Mallet, C. GPER Involvement in Inflammatory Pain. Steroids 2023, 200, 109311. [Google Scholar] [CrossRef]
- Prossnitz, E.R.; Barton, M. The G protein-coupled oestrogen receptor GPER in health and disease: An update. Nat. Rev. Endocrinol. 2023, 19, 407–424. [Google Scholar] [CrossRef]
- Wnuk, A.; Przepiórska, K.; Pietrzak, B.A.; Kajta, M. Emerging Evidence on Membrane Estrogen Receptors as Novel Therapeutic Targets for Central Nervous System Pathologies. Int. J. Mol. Sci. 2023, 24, 4043. [Google Scholar] [CrossRef] [PubMed]
- Hall, K.A.; Filardo, E.J. The G Protein-Coupled Estrogen Receptor (GPER): A Critical Therapeutic Target for Cancer. Cells 2023, 12, 2460. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Prossnitz, E.R. Targeting the G protein-coupled estrogen receptor (GPER) in obesity and diabetes. Endocr. Metab. Sci. 2021, 2, 100080. [Google Scholar] [CrossRef] [PubMed]
- Strong, A.L.; A Strong, T.; Rhodes, L.V.; A Semon, J.; Zhang, X.; Shi, Z.; Zhang, S.; Gimble, J.M.; E Burow, M.; A Bunnell, B. Obesity associated alterations in the biology of adipose stem cells mediate enhanced tumorigenesis by estrogen dependent pathways. Breast Cancer Res. 2013, 15, R102. [Google Scholar] [CrossRef] [PubMed]
- Cox-York, K.A.; Erickson, C.B.; Pereira, R.I.; Bessesen, D.H.; Van Pelt, R.E. Region-specific effects of oestradiol on adipose-derived stem cell differentiation in post-menopausal women. J. Cell. Mol. Med. 2017, 21, 677–684. [Google Scholar] [CrossRef]
- Strong, A.L.; Ohlstein, J.F.; Biagas, B.A.; Rhodes, L.V.; Pei, D.T.; Tucker, H.A.; Llamas, C.; Bowles, A.C.; Dutreil, M.F.; Zhang, S.; et al. Leptin produced by obese adipose stromal/stem cells enhances proliferation and metastasis of estrogen receptor positive breast cancers. Breast Cancer Res. 2015, 17, 112. [Google Scholar] [CrossRef] [PubMed]
- Brock, C.K.; Hebert, K.L.; Artiles, M.; Wright, M.K.; Cheng, T.; Windsor, G.O.; Nguyen, K.; Alzoubi, M.S.; Collins-Burow, B.M.; Martin, E.C.; et al. A Role for Adipocytes and Adipose Stem Cells in the Breast Tumor Microenvironment and Regenerative Medicine. Front. Physiol. 2021, 12, 751239. [Google Scholar] [CrossRef] [PubMed]
- Hoang, V.T.; Nguyen, H.-P.; Nguyen, V.N.; Hoang, D.M.; Nguyen, T.-S.T.; Thanh, L.N. “Adipose-derived mesenchymal stem cell therapy for the management of female sexual dysfunction: Literature reviews and study design of a clinical trial”. Front. Cell Dev. Biol. 2022, 10, 956274. [Google Scholar] [CrossRef] [PubMed]
- Al-Ghadban, S.; Diaz, Z.T.; Singer, H.J.; Mert, K.B.; Bunnell, B.A. Increase in Leptin and PPAR-γ Gene Expression in Lipedema Adipocytes Differentiated in vitro from Adipose-Derived Stem Cells. Cells 2020, 9, 430. [Google Scholar] [CrossRef] [PubMed]
- Kuryłowicz, A. Estrogens in Adipose Tissue Physiology and Obesity-Related Dysfunction. Biomedicines 2023, 11, 690. [Google Scholar] [CrossRef] [PubMed]
- Hetemäki, N.; Mikkola, T.S.; Tikkanen, M.J.; Wang, F.; Hämäläinen, E.; Turpeinen, U.; Haanpää, M.; Vihma, V.; Savolainen-Peltonen, H. Adipose tissue estrogen production and metabolism in premenopausalwomen. J. Steroid Biochem. Mol. Biol. 2021, 209, 105849. [Google Scholar] [CrossRef] [PubMed]
- Steiner, B.M.; Berry, D.C. The Regulation of Adipose Tissue Health by Estrogens. Front. Endocrinol. 2022, 13, 889923. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, F.; Kamble, P.G.; Hetty, S.; Fanni, G.; Vranic, M.; Sarsenbayeva, A.; Kristófi, R.; Almby, K.; Svensson, M.K.; Pereira, M.J.; et al. Role of Estrogen and Its Receptors in Adipose Tissue Glucose Metabolism in Pre- and Postmenopausal Women. J. Clin. Endocrinol. Metab. 2022, 107, e1879–e1889. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.; Chen, X.; Xing, L.; Xu, W.; Fu, X.; Zhu, J.; Mou, X.; Wang, Z.; Shu, J. Tissue-specific effects of estrogen on glycerol channel aquaporin 7 expression in an ovariectomized mouse model of menopause. Climacteric 2017, 20, 385–390. [Google Scholar] [CrossRef]
- Calis, M.; Demirtas, T.T.; Atilla, P.; Tatar, İ.; Ersoy, O.; Irmak, G.; Celik, H.H.; Cakar, A.N.; Gumusderelioglu, M.; Ozgur, F. Estrogen as a novel agent for induction of adipose-derived mesenchymal stem cells for osteogenic differentiation: In Vivo bone tissue-engineering study. Plast. Reconstr. Surg. 2014, 133, 499e–510e. [Google Scholar] [CrossRef]
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol. 2014, 12, 207–218. [Google Scholar] [CrossRef]
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning From 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7, 33. [Google Scholar] [CrossRef]
- Whitman, N.A.; Lin, Z.-W.; Kenney, R.M.; Albertini, L.; Lockett, M.R. Hypoxia differentially regulates estrogen receptor alpha in 2D and 3D culture formats. Arch. Biochem. Biophys. 2019, 671, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Bitirim, C.V.; Ozer, Z.B.; Akcali, K.C. Estrogen receptor alpha regulates the expression of adipogenic genes genetically and epige-netically in rat bone marrow-derived mesenchymal stem cells. PeerJ 2021, 9, e12071. [Google Scholar] [CrossRef] [PubMed]
- Saavedra-Peña, R.d.M.; Taylor, N.; Flannery, C.; Rodeheffer, M.S. Estradiol cycling drives female obesogenic adipocyte hyperplasia. Cell Rep. 2023, 42, 112390. [Google Scholar] [CrossRef] [PubMed]
- Shehu, A.; Albarracin, C.; Devi, Y.S.; Luther, K.; Halperin, J.; Le, J.; Mao, J.; Duan, R.W.; Frasor, J.; Gibori, G. The stimulation of HSD17B7 expression by estradiol provides a powerful feed-forward mechanism for estradiol biosynthesis in breast cancer cells. Mol. Endocrinol. 2011, 25, 754–766. [Google Scholar] [CrossRef]
- Hilborn, E.; Stål, O.; Jansson, A. Estrogen and androgen-converting enzymes 17β-hydroxysteroid dehydrogenase and their involvement in cancer: With a special focus on 17β-hydroxysteroid dehydrogenase type 1, 2, and breast cancer. Oncotarget 2017, 8, 30552–30562. [Google Scholar] [CrossRef] [PubMed]
- Cornel, K.M.C.; Kruitwagen, R.F.P.M.; Delvoux, B.; Visconti, L.; Van De Vijver, K.K.; Day, J.M.; Van Gorp, T.; Hermans, R.J.J.; Dunselman, G.A.; Romano, A. Overexpression of 17β-Hydroxysteroid Dehydrogenase Type 1 Increases the Exposure of Endometrial Cancer to 17β-Estradiol. J. Clin. Endocrinol. Metab. 2012, 97, E591–E601. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Vihma, V.; Soronen, J.; Turpeinen, U.; Hämäläinen, E.; Savolainen-Peltonen, H.; Mikkola, T.S.; Naukkarinen, J.; Pietiläinen, K.H.; Jauhiainen, M.; et al. 17β-Estradiol and estradiol fatty acyl esters and estrogen-converting enzyme expression in adipose tissue in obese men and women. J. Clin. Endocrinol. Metab. 2013, 98, 4923–4931. [Google Scholar] [CrossRef] [PubMed]
- Szafran, H.; Smielak-Korombel, W. The role of estrogens in hormonal regulation of lipid metabolism in women. Prz. Lek. 1998, 55, 266–270. [Google Scholar]
- Berger, J.J.; Barnard, R.J.; El Hafidi, M.; Pérez, I.; Zamora, J.; Soto, V.; Carvajal-Sandoval, G.; Baños, G.; Soria, A.; D’Alessandro, M.E.; et al. Effect of diet on fat cell size and hormone-sensitive lipase activity. J. Appl. Physiol. 1999, 87, 227–232. [Google Scholar] [CrossRef]
- Osuga, J.-I.; Ishibashi, S.; Oka, T.; Yagyu, H.; Tozawa, R.; Fujimoto, A.; Shionoiri, F.; Yahagi, N.; Kraemer, F.B.; Tsutsumi, O.; et al. Targeted disruption of hormone-sensitive lipase results in male sterility and adipocyte hypertrophy, but not in obesity. Proc. Natl. Acad. Sci. USA 2000, 97, 787–792. [Google Scholar] [CrossRef] [PubMed]
- Haemmerle, G.; Zimmermann, R.; Zechner, R. Letting lipids go: Hormone-sensitive lipase. Curr. Opin. Lipidol. 2003, 14, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Yeaman, S.J. Hormone-sensitive lipase-new roles for an old enzyme. Biochem. J. 2004, 379 Pt 1, 11–22. [Google Scholar] [CrossRef]
- Yi, M.; Negishi, M.; Lee, S.-J. Estrogen Sulfotransferase (SULT1E1): Its Molecular Regulation, Polymorphisms, and Clinical Perspectives. J. Pers. Med. 2021, 11, 194. [Google Scholar] [CrossRef]
- Tsunoda, Y.; Shimizu, Y.; Tsunoda, A.; Takimoto, M.; Sakamoto, M.-A.; Kusano, M. Steroid sulfatase in breast carcinoma and change of serum estrogens levels after operation. Acta Oncol. 2006, 45, 584–589. [Google Scholar] [CrossRef] [PubMed]
- Newman, S.; Purohit, A.; Ghilchik, M.; Potter, B.; Reed, M. Regulation of steroid sulphatase expression and activity in breast cancer. J. Steroid Biochem. Mol. Biol. 2000, 75, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Foster, P.A. Steroid Sulphatase and Its Inhibitors: Past, Present, and Future. Molecules 2021, 26, 2852. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Barros-Oliveira, M.d.C.; Costa-Silva, D.R.; dos Santos, A.R.; Pereira, R.O.; Soares-Júnior, J.M.; da Silva, B.B. Influence of CYP19A1 gene expression levels in women with breast cancer: A systematic review of the literature. Clinics 2021, 76, e2846. [Google Scholar] [CrossRef] [PubMed]
- Germain, D. Estrogen Carcinogenesis in Breast Cancer. Endocrinol. Metab. Clin. North Am. 2011, 40, 473–484. [Google Scholar] [CrossRef]
- Brown, K.A.; Iyengar, N.M.; Zhou, X.K.; Gucalp, A.; Subbaramaiah, K.; Wang, H.; Giri, D.D.; Morrow, M.; Falcone, D.J.; Wendel, N.K.; et al. Menopause Is a Determinant of Breast Aro-matase Expression and Its Associations With BMI, Inflammation, and Systemic Markers. J. Clin. Endocrinol. Metab. 2017, 102, 1692–1701. [Google Scholar] [CrossRef]




| Characteristics | Healthy | Lipedema |
|---|---|---|
| n | 5 | 5 |
| Sex | Female | Female |
| Age | 45 ± 6.42 | 49.8 ± 3.97 |
| BMI | 29.2 ± 2.77 | 29.9 ± 3.34 |
| Stage 2 | − | 100% |
| Name | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| ERα | GCCATGGTGGAGATCTTCGA | CCTCTCCCTGCAGATTCATCA |
| Erβ | AGAGCTCCTGGTGTGAAGCAA | GACAGCGCAGTGAGCATC |
| GPER1 | TTCCGCGAGAAGATGACCATCC | TAGTACCGCTCGTGCAGGTTGA |
| PPARG2 | AGGCGAGGGCGATCTTG | CCCATCATTAAGGAATTCATGTCATA |
| GAPDH | CGCTGAGTACGTCGTGGAGTC | GCAGGAGGCATTGCAGATGA |
| STS | GGACTGGAGTGTGGGGCAGAT | GTGCTCCCTGGTCCGATGTG |
| LIPE | AGACTTCCGCCTGGGTGCCT | CGGCGCATCGGCTCTGCTAT |
| CYP19A1 | ACTACAACCGGGTATATGGAGAA | TCGAGAGCTGTAATGATTGTGC |
| HSD17B1 | GAGCGTGGGAGGATTGATGG | AGGCTCAAGTGGACCCCAAA |
| HSD17B7 | TTGACACCATATAATGGAACAGAAG | TGATCAGAGGATTGAGAGATTCAG |
| HSD17B12 | GCCAACTTTGGATAAGCCCTCTC | AGGCAGGTTTGAGATTATCGAGC |
| SULT1E1 | TGCCACCTGAACTTCTTCCTGC | CCAGGATTTGGATGACCAGCCA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Ghadban, S.; Isern, S.U.; Herbst, K.L.; Bunnell, B.A. The Expression of Adipogenic Marker Is Significantly Increased in Estrogen-Treated Lipedema Adipocytes Differentiated from Adipose Stem Cells In Vitro. Biomedicines 2024, 12, 1042. https://doi.org/10.3390/biomedicines12051042
Al-Ghadban S, Isern SU, Herbst KL, Bunnell BA. The Expression of Adipogenic Marker Is Significantly Increased in Estrogen-Treated Lipedema Adipocytes Differentiated from Adipose Stem Cells In Vitro. Biomedicines. 2024; 12(5):1042. https://doi.org/10.3390/biomedicines12051042
Chicago/Turabian StyleAl-Ghadban, Sara, Spencer U. Isern, Karen L. Herbst, and Bruce A. Bunnell. 2024. "The Expression of Adipogenic Marker Is Significantly Increased in Estrogen-Treated Lipedema Adipocytes Differentiated from Adipose Stem Cells In Vitro" Biomedicines 12, no. 5: 1042. https://doi.org/10.3390/biomedicines12051042






