Transcriptome Analysis of Seed in Dormancy and Dormancy Release State of Epimedium koreanum Nakai
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Lamination
2.2. RNA Extraction and cDNA Library Construction
2.3. Gene Expression Analysis
2.4. Enrichment and Interaction Analyses
2.5. Transcription Factor Family Analysis
2.6. Quantitative Real-Time PCR (qRT-PCR)
3. Results
3.1. Morphology of Seed Embryos at Different Times
3.2. RNA Sequencing Quality
3.3. Differentially Expressed Genes (DEGs)
3.4. Gene Ontology (GO) Enrichment Analysis
3.5. Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analysis
3.6. Gene Expression in Plant Hormone Signal Transduction
3.7. Gene Expression in Starch and Sucrose Metabolism
3.8. Interaction Network Analysis
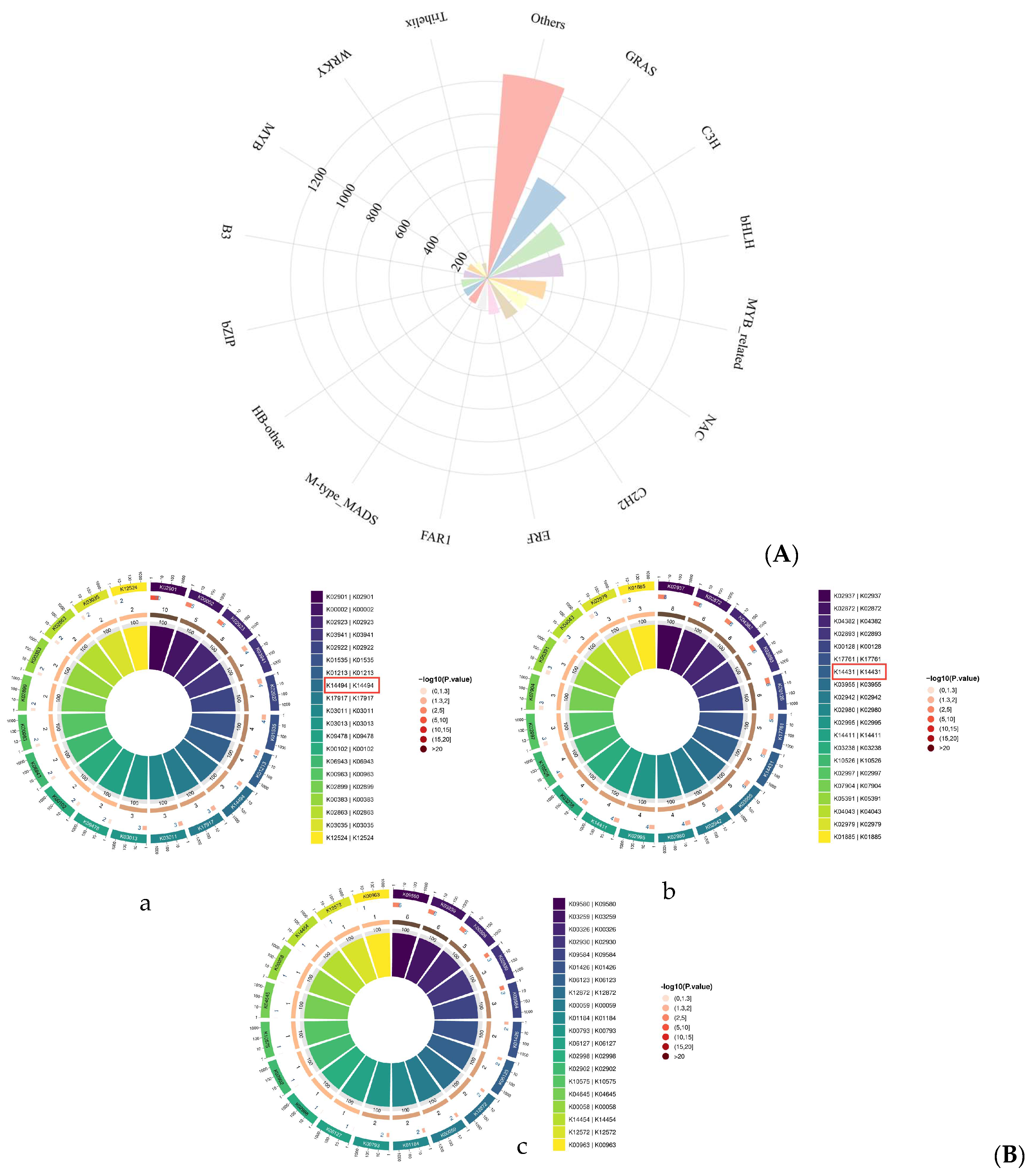
3.9. Transcription Factor Family Annotation
3.10. Analysis of Data Reliability Using Quantitative Real-Time PCR
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bewley, J.D. Seed Germination and Dormancy. Plant Cell 1997, 9, 1055–1066. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Foley, M.E. Genetic and Molecular Control of Seed Dormancy. Trends Plant Sci. 1997, 2, 384–389. [Google Scholar] [CrossRef]
- Rosbakh, S.; Baskin, C.C.; Baskin, J.M. Nikolaeva et al.’s Reference Book on Seed Dormancy and Germination. Ecology 2020, 101, e03049. [Google Scholar] [CrossRef] [PubMed]
- Baskin, J.M.; Baskin, C.C. Seed Germination Ecophysiology of Jeffersonia diphylla, a Perennial Herb of Mesic Deciduous Forests. Am. J. Bot. 1989, 76, 1073–1080. [Google Scholar] [CrossRef]
- Baskin, C.C.; Baskin, J.M.; Meyer, S.E. Seed Dormancy in the Colorado Plateau Shrub Mahonia fremontii (Berberidaceae) and Its Ecological and Evolutionary Implications. Southwest. Nat. 1993, 38, 91–99. [Google Scholar] [CrossRef]
- Buijs, G. A Perspective on Secondary Seed Dormancy in Arabidopsis thaliana. Plants 2020, 9, 749. [Google Scholar] [CrossRef] [PubMed]
- Wang, L. Studies on the Dormancy Characteristics of Rice (Oryza sativa L.) Seeds. Master’s Thesis, Hunan Normal University, Changsha, China, 2012. [Google Scholar]
- Graeber, K.; Nakabayashi, K.; Miatton, E.; Leubner-Metzger, G.; Soppe, W.J.J. Molecular Mechanisms of Seed Dormancy. Plant Cell Environ. 2012, 35, 1769–1786. [Google Scholar] [CrossRef] [PubMed]
- Baskin, J.M.; Baskin, C.C. The Great Diversity in Kinds of Seed Dormancy: A Revision of the Nikolaeva–Baskin Classification System for Primary Seed Dormancy. Seed Sci. Res. 2021, 31, 249–277. [Google Scholar] [CrossRef]
- Yu-Guo, H. Study on Embryonic Dormancy of Kalopanax septemlobus Seeds. J. Northeast. For. Univ. 1986, 1, 39–44. [Google Scholar]
- Wang, J.; Liu, P.; He, L.; Zeng, Z.; Lu, J. Factors influencing seed germimation of Epimedium koreanum Nakai. J. Shenyang Pharm. Univ. 2013, 30, 807–811. [Google Scholar] [CrossRef]
- Liu, Y.; Müller, K.; El-Kassaby, Y.A.; Kermode, A.R. Changes in Hormone Flux and Signaling in White Spruce (Picea glauca) Seeds during the Transition from Dormancy to Germination in Response to Temperature Cues. BMC Plant Biol. 2015, 15, 292. [Google Scholar] [CrossRef] [PubMed]
- National Pharmacopoeia Committee. Pharmacopoeia of the People’s Republic of China; China Medical Science and Technology Press: Beijing, China, 2020; Volume 1, p. 340. [Google Scholar]
- Yang, L.; Han, M.; Wu, J.; Han, Z.; Zhang, L. Population biomass and renewal potential of Epimedium koreanum Nakai in diffferent habitats, Linjiang, Northeast China. Acta Ecol. Sin. 2007, 27, 2251–2258. [Google Scholar]
- Rhie, Y.H.; Lee, S.Y. Seed Dormancy and Germination of Epimedium koreanum Nakai. Sci. Hortic. 2020, 272, 109600. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, L.; Wang, K.; Dong, R. Effects of Stratification Conditions and Hormone Treatments on Seed Post-Ripening of Epimedium koreanum Nakai. Mol. Plant Breed. 2024, 22, 2044–2051. [Google Scholar]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Bioinformatics: Cambridge, UK, 2010. [Google Scholar]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and Quantifying Mammalian Transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes among Gene Clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Khan, Y.; Xiong, Z.; Zhang, H.; Liu, S.; Yaseen, T.; Hui, T. Expression and Roles of GRAS Gene Family in Plant Growth, Signal Transduction, Biotic and Abiotic Stress Resistance and Symbiosis Formation—A Review. Plant Biol. 2022, 24, 404–416. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ma, C.; Xu, Y.; Wei, Q.; Imtiaz, M.; Lan, H.; Gao, S.; Cheng, L.; Wang, M.; Fei, Z.; et al. A Zinc Finger Protein Regulates Flowering Time and Abiotic Stress Tolerance in Chrysanthemum by Modulating Gibberellin Biosynthesis. Plant Cell 2014, 26, 2038–2054. [Google Scholar] [CrossRef]
- Lu, R.; Zhang, J.; Wu, Y.-W.; Wang, Y.; Zhang, J.; Zheng, Y.; Li, Y.; Li, X.-B. bHLH Transcription Factors LP1 and LP2 Regulate Longitudinal Cell Elongation. Plant Physiol. 2021, 187, 2577–2591. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Xu, R.; Li, Y. Molecular Networks of Seed Size Control in Plants. Annu. Rev. Plant Biol. 2019, 70, 435–463. [Google Scholar] [CrossRef]
- Sohn, S.-I.; Pandian, S.; Kumar, T.S.; Zoclanclounon, Y.A.B.; Muthuramalingam, P.; Shilpha, J.; Satish, L.; Ramesh, M. Seed Dormancy and Pre-Harvest Sprouting in Rice—An Updated Overview. Int. J. Mol. Sci. 2021, 22, 11804. [Google Scholar] [CrossRef] [PubMed]
- Nelson, S.K.; Steber, C.M. Gibberellin Hormone Signal Perception: Down-Regulating DELLA Repressors of Plant Growth and Development. Annu. Plant Rev. 2016, 49, 153–188. [Google Scholar]
- Song, S.; Liu, J.; Huang, H.; Wu, X.; Xu, H.; Zhang, Q.; Li, X.; Liang, J. Gibberellin hormone signal perception: Down-regulating DELLA repressors of plant growth and development. Sci. Sin. Vitae 2020, 50, 599–615. [Google Scholar]
- Finkelstein, R. Abscisic Acid Synthesis and Response. Arab. Book 2013, 11, e0166. [Google Scholar] [CrossRef]
- Kieber, J.J.; Schaller, G.E. Cytokinin Signaling in Plant Development. Development 2018, 145, dev149344. [Google Scholar] [CrossRef]
- Ma, H.; Liang, Z.; Huang, L.; Yan, C.; Kong, X. Effects of four kinds of exogenous hormones on the germination and seedling growth of Leymus chinensis. Agric. Res. Arid Areas 2008, 26, 69–73. [Google Scholar]
- Park, J.; Kim, Y.-S.; Kim, S.-G.; Jung, J.-H.; Woo, J.-C.; Park, C.-M. Integration of Auxin and Salt Signals by the NAC Transcription Factor NTM2 during Seed Germination in Arabidopsis. Plant Physiol. 2011, 156, 537–549. [Google Scholar] [CrossRef] [PubMed]
- Remington, D.L.; Vision, T.J.; Guilfoyle, T.J.; Reed, J.W. Contrasting Modes of Diversification in the Aux/IAA and ARF Gene Families. Plant Physiol. 2004, 135, 1738–1752. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin Biosynthesis by the YUCCA Flavin Monooxygenases Controls the Formation of Floral Organs and Vascular Tissues in Arabidopsis. Genes Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef] [PubMed]
- Nougué, O.; Corbi, J.; Ball, S.G.; Manicacci, D.; Tenaillon, M.I. Molecular Evolution Accompanying Functional Divergence of Duplicated Genes along the Plant Starch Biosynthesis Pathway. BMC Evol. Biol. 2014, 14, 103. [Google Scholar] [CrossRef] [PubMed]
- Keeling, P.L.; Wood, J.R.; Tyson, R.H.; Bridges, I.G. Starch Biosynthesis in Developing Wheat Grain: Evidence against the Direct Involvement of Triose Phosphates in the Metabolic Pathway. Plant Physiol. 1988, 87, 311–319. [Google Scholar] [CrossRef]
- Zhang, K.; Xia, X.; Zhang, Y.; Gan, S. An ABA-Regulated and Golgi-Localized Protein Phosphatase Controls Water Loss during Leaf Senescence in Arabidopsis. Plant J. Cell Mol. Biol. 2012, 69, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Leung, J.; Merlot, S.; Giraudat, J. The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 Genes Encode Homologous Protein Phosphatases 2C Involved in Abscisic Acid Signal Transduction. Plant Cell 1997, 9, 759–771. [Google Scholar] [CrossRef]
- Ruan, Y.-L. Sucrose Metabolism: Gateway to Diverse Carbon Use and Sugar Signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef]
- Coleman, H.D.; Yan, J.; Mansfield, S.D. Sucrose Synthase Affects Carbon Partitioning to Increase Cellulose Production and Altered Cell Wall Ultrastructure. Proc. Natl. Acad. Sci. USA 2009, 106, 13118–13123. [Google Scholar] [CrossRef]
- Xu, S.-M.; Brill, E.; Llewellyn, D.J.; Furbank, R.T.; Ruan, Y.-L. Overexpression of a Potato Sucrose Synthase Gene in Cotton Accelerates Leaf Expansion, Reduces Seed Abortion, and Enhances Fiber Production. Mol. Plant 2012, 5, 430–441. [Google Scholar] [CrossRef]
- Winter, H.; Huber, S.C. Regulation of Sucrose Metabolism in Higher Plants: Localization and Regulation of Activity of Key Enzymes. Crit. Rev. Biochem. Mol. Biol. 2000, 35, 253–289. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.-K.; Chang, C. Arabidopsis RGL1 Encodes a Negative Regulator of Gibberellin Responses. Plant Cell 2002, 14, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.P.; Piskurewicz, U.; Turečková, V.; Strnad, M.; Lopez-Molina, L. A Seed Coat Bedding Assay Shows That RGL2-Dependent Release of Abscisic Acid by the Endosperm Controls Embryo Growth in Arabidopsis Dormant Seeds. Proc. Natl. Acad. Sci. USA 2010, 107, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Xing, M.-Q.; Chen, S.-H.; Zhang, X.-F.; Xue, H.-W. Rice OsGA2ox9 Regulates Seed GA Metabolism and Dormancy. Plant Biotechnol. J. 2023, 21, 2411–2413. [Google Scholar] [CrossRef] [PubMed]
- Hou, M. Cloning, Transformation and Functional Analysis of GRAS Transcription Factor Family Genes in Rice. Master’s Thesis, Jilin University, Changchun, China, 2013. [Google Scholar]
- Liu, Y.; Li, S.; Wang, Y.; Yang, J.; Zhou, W.; Shen, Y. Research progress on the functions of GRAS transcription factors in medical plants. J. Zhejiang A&F Univ. 2019, 36, 1233–1240. [Google Scholar]
- Sun, X.; Jones, W.T.; Rikkerink, E.H.A. GRAS Proteins: The Versatile Roles of Intrinsically Disordered Proteins in Plant Signalling. Biochem. J. 2012, 442, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Engstrom, E.M. HAM Proteins Promote Organ Indeterminacy. Plant Signal. Behav. 2012, 7, 227–234. [Google Scholar] [CrossRef]
- Li, Z.; Thomas, T.L. PEI1, an Embryo-Specific Zinc Finger Protein Gene Required for Heart-Stage Embryo Formation in Arabidopsis. Plant Cell 1998, 10, 383–398. [Google Scholar] [CrossRef]
- Li, J.; Jia, D.; Chen, X. HUA1, a Regulator of Stamen and Carpel Identities in Arabidopsis, Codes for a Nuclear RNA Binding Protein. Plant Cell 2001, 13, 2269–2281. [Google Scholar] [CrossRef]
- Cheng, Y.; Kato, N.; Wang, W.; Li, J.; Chen, X. Two RNA Binding Proteins, HEN4 and HUA1, Act in the Processing of AGAMOUS Pre-mRNA in Arabidopsis thaliana. Dev. Cell 2003, 4, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Jeon, S.J.; Hwang, S.M.; Hong, J.C.; Bahk, J.D. The C3H-Type Zinc Finger Protein GDS1/C3H42 Is a Nuclear-Speckle-Localized Protein That Is Essential for Normal Growth and Development in Arabidopsis. Plant Sci. 2016, 250, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Jia, J.; Yan, X.; Shi, H.; Han, Y. Arabidopsis KHZ1 and KHZ2, Two Novel Non-Tandem CCCH Zinc-Finger and K-Homolog Domain Proteins, Have Redundant Roles in the Regulation of Flowering and Senescence. Plant Mol. Biol. 2017, 95, 549–565. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Xu, Z.; Cao, S.; Chen, K.; Li, S.; Liu, X.; Gao, C.; Zhang, B.; Zhou, Y. An Uncanonical CCCH-Tandem Zinc-Finger Protein Represses Secondary Wall Synthesis and Controls Mechanical Strength in Rice. Mol. Plant 2018, 11, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Kong, Z.; Li, M.; Yang, W.; Xu, W.; Xue, Y. A Novel Nuclear-Localized CCCH-Type Zinc Finger Protein, OsDOS, Is Involved in Delaying Leaf Senescence in Rice. Plant Physiol. 2006, 141, 1376–1388. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xu, Y.; Zhang, C.; Ma, Q.; Joo, S.-H.; Kim, S.-K.; Xu, Z.; Chong, K. OsLIC, a Novel CCCH-Type Zinc Finger Protein with Transcription Activation, Mediates Rice Architecture via Brassinosteroids Signaling. PLoS ONE 2008, 3, e3521. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, F.; Zhou, J.; Fan, Z.; Chen, F.; Ma, H.; Xie, X. Overexpression of a Phytochrome-Regulated Tandem Zinc Finger Protein Gene, OsTZF1, Confers Hypersensitivity to ABA and Hyposensitivity to Red Light and Far-Red Light in Rice Seedlings. Plant Cell Rep. 2012, 31, 1333–1343. [Google Scholar] [CrossRef]
- Li, P.; Xiang, Q.; Dong, X.; Wang, Y. Characterizing Seed Dormancy in Epimedium brevicornu Maxim.: Development of Novel Chill Models and Determination of Dormancy Release Mechanisms by Transcriptomics. BMC Plant Biol. 2023. [Google Scholar] [CrossRef]
- Ma, Y.; Chen, X.; Guo, B. Identification of Genes Involved in Metabolism and Signalling of Abscisic Acid and Gibberellins during Epimedium pseudowushanense B.L. Guo Seed Morphophysiological Dormancy. Plant Cell Rep. 2018, 37, 1061–1075. [Google Scholar] [CrossRef]









| Sample | Raw Reads | Raw Bases | Valid Reads | Valid Bases | Valid% | Q20% | Q30% | GC% |
|---|---|---|---|---|---|---|---|---|
| NS_1 | 41,957,680 | 6.29 G | 41,393,464 | 5.77 G | 98.66 | 97.74 | 93.16 | 47.41 |
| NS_2 | 40,797,168 | 6.12 G | 40,226,414 | 5.61 G | 98.60 | 97.76 | 93.21 | 47.46 |
| NS_3 | 40,803,704 | 6.12 G | 40,229,626 | 5.61 G | 98.59 | 97.75 | 93.19 | 47.42 |
| CS_1 | 40,468,880 | 6.07 G | 39,924,266 | 5.57 G | 98.65 | 97.79 | 93.46 | 55.40 |
| CS_2 | 39,476,264 | 5.92 G | 38,970,190 | 5.43 G | 98.72 | 97.86 | 93.65 | 55.45 |
| CS_3 | 41,336,382 | 6.20 G | 40,794,118 | 5.69 G | 98.69 | 97.85 | 93.63 | 55.52 |
| ND_1 | 43,005,770 | 6.45 G | 42,169,380 | 5.88 G | 98.06 | 97.91 | 93.63 | 52.44 |
| ND_2 | 40,732,900 | 6.11 G | 40,066,156 | 5.58 G | 98.36 | 97.79 | 93.36 | 50.43 |
| ND_3 | 44,456,686 | 6.67 G | 43,867,368 | 6.13 G | 98.67 | 98.10 | 94.14 | 52.56 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Wu, F.; Yu, J.; Zhang, Z.; Huang, X.; Hou, H.; Yang, L. Transcriptome Analysis of Seed in Dormancy and Dormancy Release State of Epimedium koreanum Nakai. Agronomy 2024, 14, 1037. https://doi.org/10.3390/agronomy14051037
Zhang Y, Wu F, Yu J, Zhang Z, Huang X, Hou H, Yang L. Transcriptome Analysis of Seed in Dormancy and Dormancy Release State of Epimedium koreanum Nakai. Agronomy. 2024; 14(5):1037. https://doi.org/10.3390/agronomy14051037
Chicago/Turabian StyleZhang, Yonggang, Feng Wu, Jingjing Yu, Zhiqiang Zhang, Xiangdi Huang, Huiling Hou, and Limin Yang. 2024. "Transcriptome Analysis of Seed in Dormancy and Dormancy Release State of Epimedium koreanum Nakai" Agronomy 14, no. 5: 1037. https://doi.org/10.3390/agronomy14051037




