Single-Cell Transcriptomic Profiling Unveils Dynamic Immune Cell Responses during Haemonchus contortus Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Fecal Egg Count Detection
2.3. Quantitative Real-Time PCR
2.4. Single-Cell RNA-Seq Library Preparation and Sequencing
2.5. Single-Cell RNA-Seq Data Analysis
2.6. Quality Control, Dimension Reduction, and Cell Clustering
2.7. Cell Subpopulation Annotation
2.8. Cell Cycle Analysis
2.9. Gene Differential Expression Analysis
2.10. SCENIC Analysis
2.11. Cell Communication Analysis
3. Results
3.1. Parasite Load and Immune Response in H. contortus-Infected Goats
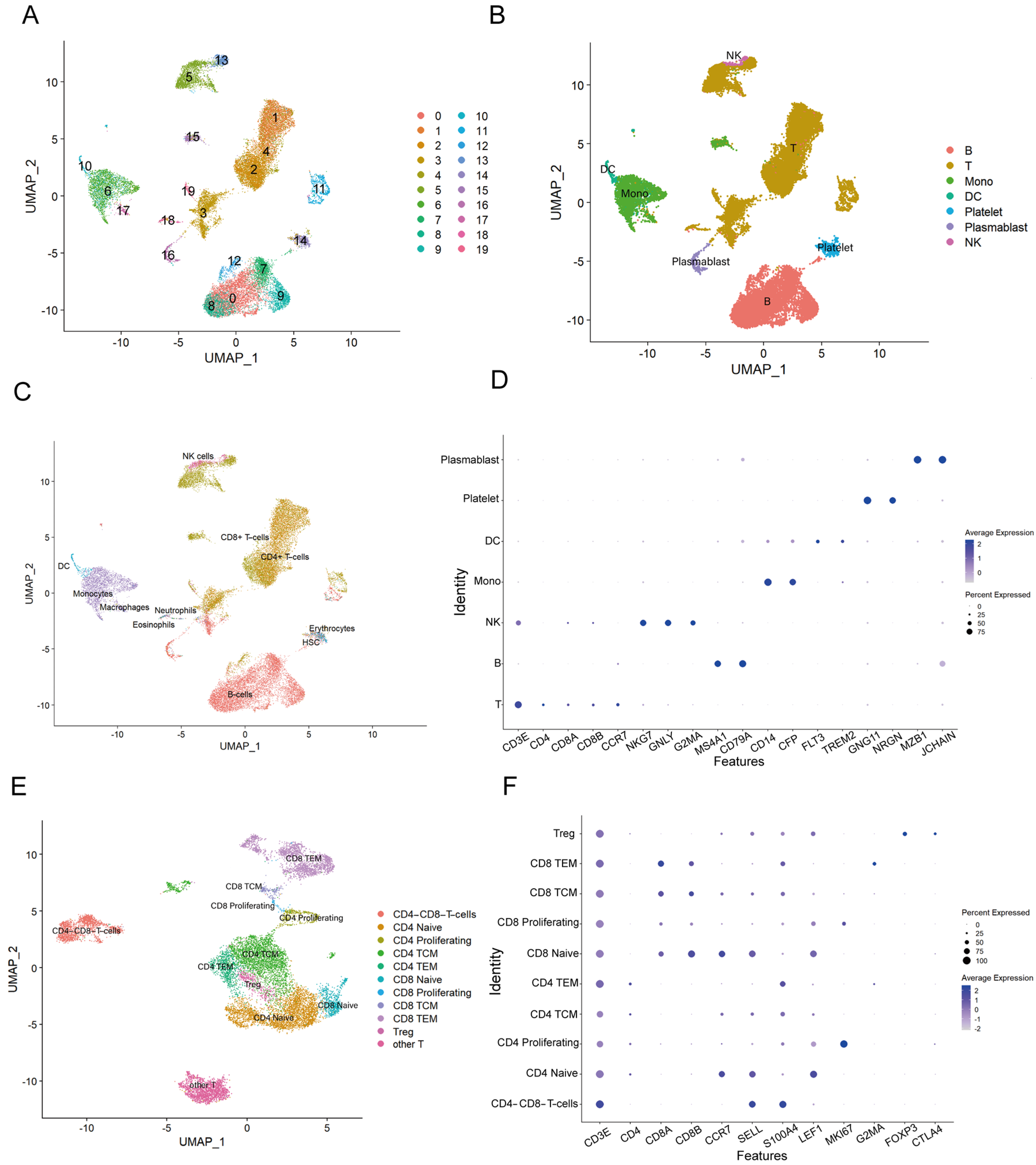
3.2. Annotation of Subpopulations in Goat PBMCs
3.3. Infection with H. contortus Alters the Gene Expression and Proportions of PBMCs
3.4. Infection with H. contortus Enhances Intercellular Communication in PBMCs
3.5. Transcriptional Factor Activation during H. contortus Infection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Flay, K.J.; Hill, F.I.; Muguiro, D.H. A Review: Haemonchus contortus Infection in Pasture-Based Sheep Production Systems, with a Focus on the Pathogenesis of Anaemia and Changes in Haematological Parameters. Animals 2022, 12, 1238. [Google Scholar] [CrossRef]
- Alam, M.B.B.; Omar, A.I.; Faruque, M.O.; Notter, D.R.; Periasamy, K.; Mondal, M.M.H.; Sarder, M.J.U.; Shamsuddin, M.; Cao, J.H.; Du, X.Y.; et al. Single nucleotide polymorphisms in candidate genes are significantly associated with resistance to Haemonchus contortus infection in goats. J. Anim. Sci. Biotechnol. 2019, 10, 30. [Google Scholar] [CrossRef]
- Bhuiyan, A.A.; Li, J.; Wu, Z.; Ni, P.; Adetula, A.A.; Wang, H.; Zhang, C.; Tang, X.; Bhuyan, A.A.; Zhao, S.; et al. Exploring the Genetic Resistance to Gastrointestinal Nematodes Infection in Goat Using RNA-Sequencing. Int. J. Mol. Sci. 2017, 18, 751. [Google Scholar] [CrossRef]
- Castagna, F.; Bava, R.; Palma, E.; Morittu, V.; Spina, A.; Ceniti, C.; Lupia, C.; Cringoli, G.; Rinaldi, L.; Bosco, A.; et al. Effect of pomegranate (Punica granatum) anthelmintic treatment on milk production in dairy sheep naturally infected with gastrointestinal nematodes. Front. Vet. Sci. 2024, 11, 1347151. [Google Scholar] [CrossRef]
- Saddiqi, H.A.; Jabbar, A.; Sarwar, M.; Iqbal, Z.; Muhammad, G.; Nisa, M.; Shahzad, A. Small ruminant resistance against gastrointestinal nematodes: A case of Haemonchus contortus. Parasitol. Res. 2011, 109, 1483–1500. [Google Scholar] [CrossRef]
- Hoffmann, J.A.; Kafatos, F.C.; Janeway, C.A.; Ezekowitz, R.A.B. Phylogenetic perspectives in innate immunity. Science 1999, 284, 1313–1318. [Google Scholar] [CrossRef]
- Sakai, K.; Sakurai, T.; De Velasco, M.A.; Nagai, T.; Chikugo, T.; Ueshima, K.; Kura, Y.; Takahama, T.; Hayashi, H.; Nakagawa, K.; et al. Intestinal Microbiota and Gene Expression Reveal Similarity and Dissimilarity between Immune-Mediated Colitis and Ulcerative Colitis. Front. Oncol. 2021, 11, 763468. [Google Scholar] [CrossRef]
- Finney, C.A.M.; Lu, Z.; LeBourhis, L.; Philpott, D.J.; Kain, K.C. Disruption of Nod-like Receptors Alters Inflammatory Response to Infection but Does Not Confer Protection in Experimental Cerebral Malaria. Am. J. Trop. Med. Hyg. 2009, 80, 718–722. [Google Scholar] [CrossRef]
- Goodridge, H.S.; Marshall, F.A.; Else, K.J.; Houston, K.M.; Egan, C.; Al-Riyami, L.; Liew, F.Y.; Harnett, W.; Harnett, M.M. Immunomodulation via novel use of TLR4 by the filarial nematode phosphorylcholine-containing secreted product, ES-62. J. Immunol. 2005, 174, 284–293. [Google Scholar] [CrossRef]
- Dibo, N.; Liu, X.S.; Chang, Y.F.; Huang, S.Q.; Wu, X. Pattern recognition receptor signaling and innate immune responses to schistosome infection. Front. Cell. Infect. Microbiol. 2022, 12, 1040270. [Google Scholar] [CrossRef]
- Wen, Z.H.; Zhang, Y.; Feng, J.J.; Aimulajiang, K.; Aleem, M.T.; Lu, M.M.; Xu, L.X.; Song, X.K.; Li, X.R.; Yan, R.F. Excretory/secretory proteins inhibit host immune responses by downregulating the TLR4/NF-κB/MAPKs signaling pathway: A possible mechanism of immune evasion in parasitic nematode Haemonchus contortus. Front. Immunol. 2022, 13, 1013159. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Hu, D.D.; Wang, C.Y.; Tang, X.M.; Du, M.Z.; Gu, X.L.; Suo, J.X.; Hu, M.; Fang, R.; Zhu, X.Q.; et al. Transcriptional profiling of innate immune responses in sheep PBMCs induced by Haemonchus contortus soluble extracts. Parasite Vector 2019, 12, 182. [Google Scholar] [CrossRef] [PubMed]
- Toscano, J.H.B.; Okino, C.H.; dos Santos, I.B.; Giraldelo, L.A.; von Haehling, M.B.; Esteves, S.N.; Chagas, A.C.D. Innate Immune Responses Associated with Resistance against Haemonchus contortus in Morada Nova Sheep. J. Immunol. Res. 2019, 2019, 3562672. [Google Scholar] [CrossRef]
- Kalia, I.; Anand, R.; Quadiri, A.; Bhattacharya, S.; Sahoo, B.; Singh, A.P. Plasmodium berghei-Released Factor, PbTIP, Modulates the Host Innate Immune Responses. Front. Immunol. 2021, 12, 699887. [Google Scholar] [CrossRef]
- Lu, M.M.; Tian, X.W.; Tian, A.L.; Li, C.; Yan, R.F.; Xu, L.X.; Song, X.K.; Li, X.R. A Novel α/β Hydrolase Domain Protein Derived from Haemonchus contortus Acts at the Parasite-Host Interface. Front. Immunol. 2020, 11, 1388. [Google Scholar] [CrossRef]
- Gadahi, J.A.; Li, B.J.; Ehsan, M.; Wang, S.; Zhang, Z.C.; Wang, Y.J.; Hasan, M.W.; Yan, R.F.; Song, X.K.; Xu, L.X.; et al. Recombinant Haemonchus contortus 24 kDa excretory/secretory protein (rHcES-24) modulate the immune functions of goat PBMCs. Oncotarget 2016, 7, 83926–83937. [Google Scholar] [CrossRef]
- Li, Z.B.; Sun, C.C.; Wang, F.; Wang, X.R.; Zhu, J.C.; Luo, L.H.; Ding, X.N.; Zhang, Y.N.; Ding, P.W.; Wang, H.Y.; et al. Molecular mechanisms governing circulating immune cell heterogeneity across different species revealed by single-cell sequencing. Clin. Transl. Med. 2022, 12, e689. [Google Scholar] [CrossRef]
- McGinnis, C.S.; Murrow, L.M.; Gartner, Z.J. DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst. 2019, 8, 329–337.e4. [Google Scholar] [CrossRef]
- Korsunsky, I.; Millard, N.; Fan, J.; Slowikowski, K.; Zhang, F.; Wei, K.; Baglaenko, Y.; Brenner, M.; Loh, P.R.; Raychaudhuri, S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 2019, 16, 1289–1296. [Google Scholar] [CrossRef]
- Franzén, O.; Gan, L.M.; Björkegren, J.L.M. PanglaoDB: A web server for exploration of mouse and human single-cell RNA sequencing data. Database 2019, 2019, baz046. [Google Scholar] [CrossRef]
- Hu, C.X.; Li, T.Y.; Xu, Y.Q.; Zhang, X.X.; Li, F.; Bai, J.; Chen, J.; Jiang, W.Q.; Yang, K.Y.; Ou, Q.; et al. CellMarker 2.0: An updated database of manually curated cell markers in human/mouse and web tools based on scRNA-seq data. Nucleic Acids Res. 2023, 51, D870–D876. [Google Scholar] [CrossRef]
- Aran, D.; Looney, A.P.; Liu, L.Q.; Wu, E.; Fong, V.; Hsu, A.; Chak, S.; Naikawadi, R.P.; Wolters, P.J.; Abate, A.R.; et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019, 20, 163–172. [Google Scholar] [CrossRef]
- Aibar, S.; González-Blas, C.B.; Moerman, T.; Van, A.H.T.; Imrichova, H.; Hulselmans, G.; Rambow, F.; Marine, J.C.; Geurts, P.; Aerts, J.; et al. SCENIC: Single-cell regulatory network inference and clustering. Nat. Methods 2017, 14, 1083–1086. [Google Scholar] [CrossRef]
- Jin, S.Q.; Guerrero-Juarez, C.F.; Zhang, L.H.; Chang, I.; Ramos, R.; Kuan, C.H.; Myung, P.; Plikus, M.V.; Nie, Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021, 12, 1088. [Google Scholar] [CrossRef]
- Bricarello, P.A.; Zaros, L.G.; Coutinho, L.L.; Rocha, R.A.; Silva, M.B.; Kooyman, F.N.J.; De Vries, E.; Yatsuda, A.P.; Amarante, A.F.T. Immunological responses and cytokine gene expression analysis to Cooperia punctata infections in resistant and susceptible Nelore cattle. Vet. Parasitol. 2008, 155, 95–103. [Google Scholar] [CrossRef]
- Balic, A.; Bowles, V.M.; Meeusen, E.N.T. Cellular profiles in the abomasal mucosa and lymph node during primary infection with Haemonchus contortus in sheep. Vet. Immunol. Immunopathol. 2000, 75, 109–120. [Google Scholar] [CrossRef]
- Halliday, A.M.; Morrison, W.I.; Smith, W.D. Kinetics of the local cellular response in the gastric lymph of immune and susceptible sheep to infection with Teladorsagia circumcincta. Parasite Immunol. 2009, 31, 402–411. [Google Scholar] [CrossRef]
- Jabeen, R.; Goswami, R.; Awe, O.; Kulkarni, A.; Nguyen, E.T.; Attenasio, A.; Walsh, D.; Olson, M.R.; Kim, M.H.; Tepper, R.S.; et al. Th9 cell development requires a BATF-regulated transcriptional network. J. Clin. Investig. 2013, 123, 4641–4653. [Google Scholar] [CrossRef]
- Nerlov, C. The C/EBP family of transcription factors: A paradigm for interaction between gene expression and proliferation control. Trends Cell Biol. 2007, 17, 318–324. [Google Scholar] [CrossRef]
- Tamura, A.; Hirai, H.; Yokota, A.; Kamio, N.; Sato, A.; Shoji, T.; Kashiwagi, T.; Torikoshi, Y.; Miura, Y.; Tenen, D.G.; et al. C/EBPβ is required for survival of Ly6C monocytes. Blood 2017, 130, 1809–1818. [Google Scholar] [CrossRef]
- Estrada-Reyes, Z.M.; López-Reyes, A.G.; Lagunas-Martínez, A.; Ramírez-Vargas, G.; Olazarán-Jenkins, S.; Hernández-Romano, J.; Mendoza-De-Gives, P.; López-Arellano, M.E. Relative expression analysis of IL-5 and IL-6 genes in tropical sheep breed Pelibuey infected with Haemonchus contortus. Parasite Immunol. 2015, 37, 446–452. [Google Scholar] [CrossRef]
- Liu, A.Y.; Dwyer, D.F.; Jones, T.G.; Bankova, L.G.; Shen, S.L.; Katz, H.R.; Austen, K.F.; Gurish, M.F. Mast Cells Recruited to Mesenteric Lymph Nodes during Helminth Infection Remain Hypogranular and Produce IL-4 and IL-6. J. Immunol. 2013, 190, 1758–1766. [Google Scholar] [CrossRef]
- Moro, K.; Yamada, T.; Tanabe, M.; Takeuchi, T.; Ikawa, T.; Kawamoto, H.; Furusawa, J.; Ohtani, M.; Fujii, H.; Koyasu, S. Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature 2010, 463, 540–544. [Google Scholar] [CrossRef]
- Prudente, M.F.D.; Crespo, A.D.C.; Carvalhaes, M.S. Lagochilascaris minor: Antibody production in experimentally infected mice. Rev. Soc. Bras. Med. Tro 2009, 42, 325–328. [Google Scholar] [CrossRef]
- Han, S.H.; Chae, D.S.; Kim, S.W. Dual CXCR4/IL-10 Gene-Edited Human Amniotic Mesenchymal Stem Cells Exhibit Robust Therapeutic Properties in Chronic Wound Healing. Int. J. Mol. Sci. 2022, 23, 15338. [Google Scholar] [CrossRef]
- Shen, J.; Xu, L.; Liu, Z.; Li, N.; Wang, L.F.; Lv, Z.Y.; Fung, M.C.; Wu, Z.D.; Sun, X. Gene expression profile of LPS-stimulated dendritic cells induced by a recombinant Sj16 (rSj16) derived from Schistosoma japonicum. Parasitol. Res. 2014, 113, 3073–3083. [Google Scholar] [CrossRef]
- Buchmann, K. Evolution of innate immunity: Clues from invertebrates via fish to mammals. Front. Immunol. 2014, 5, 459. [Google Scholar] [CrossRef]
- Park, S.H.; Rhee, J.; Kim, S.K.; Kang, J.A.; Kwak, J.S.; Son, Y.O.; Choi, W.S.; Park, S.G.; Chun, J.S. BATF regulates collagen-induced arthritis by regulating T helper cell differentiation. Arthritis Res. Ther. 2018, 20, 161. [Google Scholar] [CrossRef]
- Schraml, B.U.; Hildner, K.; Ise, W.; Lee, W.L.; Smith, W.A.E.; Solomon, B.; Sahota, G.; Sim, J.; Mukasa, R.; Cemerski, S.; et al. The AP-1 transcription factor Batf controls TH17 differentiation. Nature 2009, 460, 405–409. [Google Scholar] [CrossRef]
- Godec, J.; Cowley, G.S.; Barnitz, R.A.; Alkan, O.; Root, D.E.; Sharpe, A.H.; Haining, W.N. Inducible RNAi in vivo reveals that the transcription factor BATF is required to initiate but not maintain CD8+ T-cell effector differentiation. Proc. Natl. Acad. Sci. USA 2015, 112, 512–517. [Google Scholar] [CrossRef]
- Takahashi, A.; Iwabuchi, K.; Suzuki, M.; Ogasawara, K.; Nishihira, J.; Onoé, K. Antisense macrophage migration inhibitory factor (MIF) prevents anti-IgM mediated growth arrest and apoptosis of a murine B cell line by regulating cell cycle progression. Microbiol. Immunol. 1999, 43, 61–67. [Google Scholar] [CrossRef]
- Bao, K.; Carr, T.; Wu, J.X.; Barclay, W.; Jin, J.X.; Ciofani, M.; Reinhardt, R.L. BATF Modulates the Th2 Locus Control Region and Regulates CD4 T Cell Fate during Antihelminth Immunity. J. Immunol. 2016, 197, 4371–4381. [Google Scholar] [CrossRef]
- Bacher, M.; Metz, C.N.; Calandra, T.; Mayer, K.; Chesney, J.; Lohoff, M.; Gemsa, D.; Donnelly, T.; Bucala, R. An essential regulatory role for macrophage migration inhibitory factor in T-cell activation. Proc. Natl. Acad. Sci. USA 1996, 93, 7849–7854. [Google Scholar] [CrossRef]
- Calandra, T.; Bernhagen, J.; Mitchell, R.A.; Bucala, R. The macrophage is an important and previously unrecognized source of macrophage migration inhibitory factor. J. Exp. Med. 1994, 179, 1895–1902. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.; Bernhagen, J. Revisiting the secretion mechanism(s) of macrophage migration inhibitory factor-welcome to the “UPS club”. Immunol. Cell Biol. 2020, 98, 704–708. [Google Scholar] [CrossRef]
- Calandra, T.; Roger, T. Macrophage migration inhibitory factor: A regulator of innate immunity. Nat. Rev. Immunol. 2003, 3, 791–800. [Google Scholar] [CrossRef]
- Varyani, F.; Löser, S.; Filbey, K.J.; Harcus, Y.; Drurey, C.; Poveda, M.C.; Rasid, O.; White, M.P.J.; Smyth, D.J.; Gerbe, F.; et al. The IL-25-dependent tuft cell circuit driven by intestinal helminths requires macrophage migration inhibitory factor (MIF). Mucosal Immunol. 2022, 15, 1243–1256. [Google Scholar] [CrossRef]
- Marcon, C.F.; Ferreira, P.T.M.; Franco, P.S.; Ribeiro, M.; Silva, R.J.; Sousa, R.A.P.; Oliveira, C.J.F.; Rodrigues, V.; Gomes, M.L.M.; Chica, J.E.L.; et al. Macrophage migration inhibitory factor (MIF) and pregnancy may impact the balance of intestinal cytokines and the development of intestinal pathology caused by Toxoplasma gondii infection. Cytokine 2020, 136, 155283. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Jin, Z.; Kong, M.; Yan, Z.; Fu, L.; Du, X. Single-Cell Transcriptomic Profiling Unveils Dynamic Immune Cell Responses during Haemonchus contortus Infection. Cells 2024, 13, 842. https://doi.org/10.3390/cells13100842
Wang W, Jin Z, Kong M, Yan Z, Fu L, Du X. Single-Cell Transcriptomic Profiling Unveils Dynamic Immune Cell Responses during Haemonchus contortus Infection. Cells. 2024; 13(10):842. https://doi.org/10.3390/cells13100842
Chicago/Turabian StyleWang, Wenxuan, Zhe Jin, Mei Kong, Zhuofan Yan, Liangliang Fu, and Xiaoyong Du. 2024. "Single-Cell Transcriptomic Profiling Unveils Dynamic Immune Cell Responses during Haemonchus contortus Infection" Cells 13, no. 10: 842. https://doi.org/10.3390/cells13100842



