Human-Induced Range Expansions Result in a Recent Hybrid Zone between Sister Species of Ducks
Abstract
:1. Introduction
2. Methods
2.1. Sample Collection and DNA Extraction
2.2. Mitochondrial DNA Sequencing and Analyses
2.3. Nuclear DNA ddRADseq Library Preparation, Sequencing, and Bioinformatics
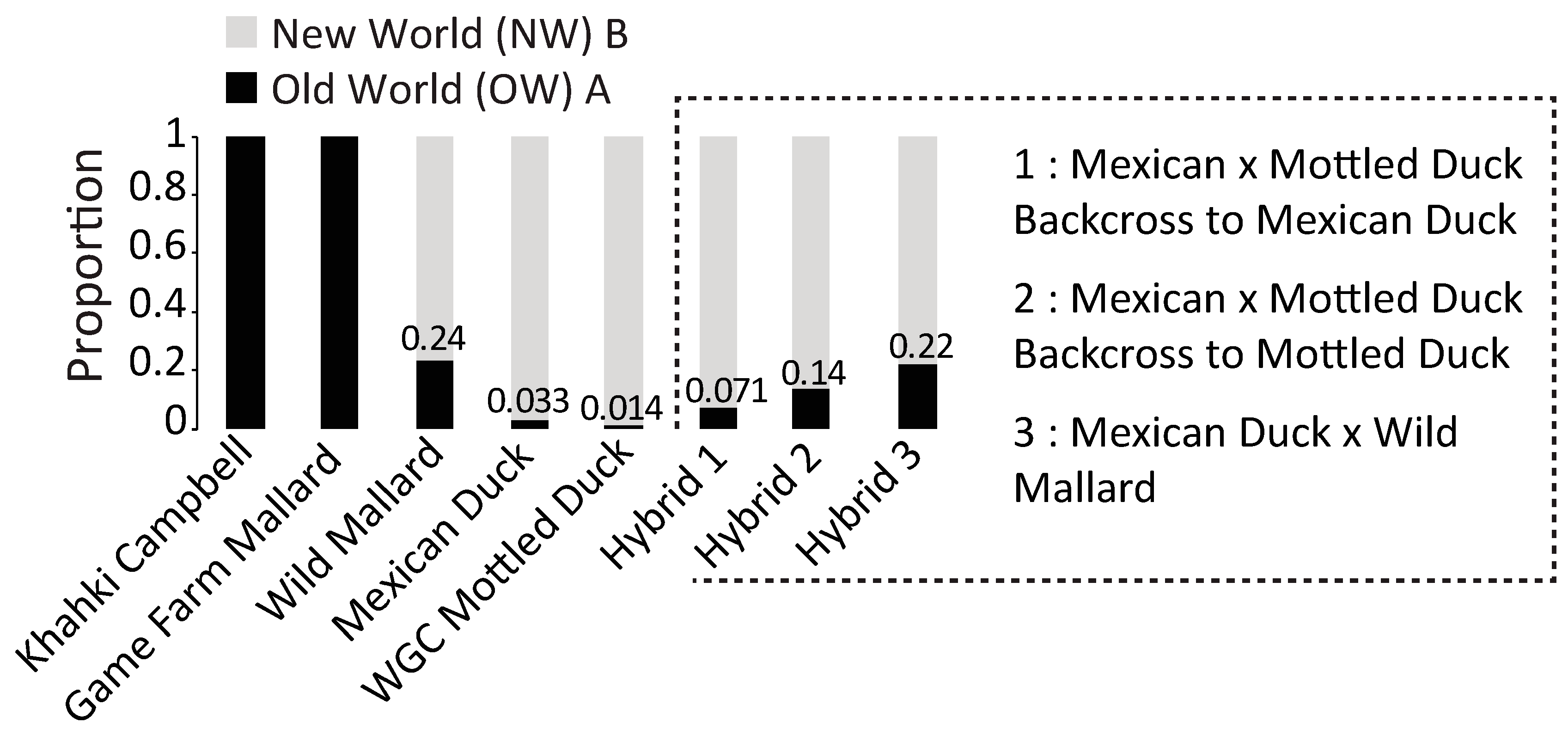
2.4. Nuclear Analyses
3. Results
3.1. Mitochondrial DNA
3.2. Nuclear DNA
4. Discussion
Conservation Considerations in the Anthropocene
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Taylor, S.A.; White, T.A.; Hochachka, W.M.; Ferretti, V.; Curry, R.L.; Lovette, I. Climate-mediated movement of an avian hybrid zone. Curr. Biol. 2014, 24, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Crispo, E.; Moore, J.; Lee-Yaw, J.A.; Gray, S.M.; Haller, B.C. Broken barriers: Human-induced changes to gene flow and introgression in animals. BioEssays 2011, 33, 508–518. [Google Scholar] [CrossRef]
- Walther, G.-R.; Post, E.; Convey, P.; Menzel, A.; Parmesan, C.; Beebee, T.J.C.; Fromentin, J.-M.; Hoegh-Guldberg, O.; Bairlein, F. Ecological responses to recent climate change. Nature 2002, 416, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, K.V.; Dokter, A.M.; Blancher, P.J.; Sauer, J.R.; Smith, A.C.; Smith, P.A.; Stanton, J.C.; Panjabi, A.; Helft, L.; Parr, M.; et al. Decline of the North American avifauna. Science 2019, 366, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Hitch, A.T.; Leberg, P.L. Breeding distributions of North American bird species moving north as a result of climate change. Conserv. Biol. 2005, 21, 534–539. [Google Scholar] [CrossRef] [PubMed]
- Trautmann, S. Climate change impacts on bird species. In Bird Species: How They Arise, Modify and Vanish; Springer: Berlin/Heidelberg, Germany, 2018; pp. 217–234. [Google Scholar]
- Hernández, F.; Brown, J.I.; Kaminski, M.; Harvey, M.G.; Lavretsky, P. Genomic Evidence for Rare Hybridization and Large Demographic Changes in the Evolutionary Histories of Four North American Dove Species. Animals 2021, 11, 2677. [Google Scholar] [CrossRef] [PubMed]
- Ottenburghs, J. The genic view of hybridization in the Anthropocene. Evol. Appl. 2021, 14, 2342–2360. [Google Scholar] [CrossRef] [PubMed]
- Larson, E.L.; Tinghitella, R.M.; Taylor, S.A. Insect hybridization and climate change. Front. Ecol. Evol. 2019, 7, 348. [Google Scholar] [CrossRef]
- Hohenlohe, P.A.; Funk, W.C.; Rajora, O.P. Population genomics for wildlife conservation and management. Mol. Ecol. 2021, 30, 62–82. [Google Scholar] [CrossRef]
- Lavretsky, P.; McCracken, K.G.; Peters, J.L. Phylogenetics of a recent radiation in the mallards and allies (Aves: Anas): Inferences from a genomic transect and the multispecies coalescent. Mol. Phylogenetics Evol. 2014, 70, 402–411. [Google Scholar] [CrossRef]
- Lavretsky, P.; Mohl, J.E.; Söderquist, P.; Kraus, R.H.S.; Schummer, M.L.; Brown, J.I. The meaning of wild: Genetic and adaptive consequences from large-scale releases of domestic mallards. Commun. Biol. 2023, 6, 819. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.I.; Harrigan, R.J.; Lavretsky, P. Evolutionary and Ecological Drivers of Local Adaptation and Speciation in a North American Avian Species Complex. Mol. Ecol. 2022, 31, 2578–2593. [Google Scholar] [CrossRef]
- Lavretsky, P.; McInerney, N.R.; Mohl, J.E.; Brown, J.I.; James, H.F.; McCracken, K.G.; Fleischer, R.C. Assessing changes in genomic divergence following a century of human-mediated secondary contact among wild and captive-bred ducks. Mol. Ecol. 2020, 29, 578–595. [Google Scholar] [CrossRef]
- Lavretsky, P.; Janzen, T.; McCracken, K.G. Identifying hybrids & the genomics of hybridization: Mallards & American black ducks of eastern North America. Ecol. Evol. 2019, 9, 3470–3490. [Google Scholar] [CrossRef] [PubMed]
- Lavretsky, P.; DaCosta, J.M.; Sorenson, M.D.; McCracken, K.G.; Peters, J.L. ddRAD-seq data reveal significant genome-wide population structure and divergent genomic regions that distinguish the mallard and close relatives in North America. Mol. Ecol. 2019, 28, 2594–2609. [Google Scholar] [CrossRef] [PubMed]
- Lavretsky, P.; Dacosta, J.M.; Hernández-Baños, B.E.; Engilis, A.; Sorenson, M.D.; Peters, J.L. Speciation genomics and a role for the Z chromosome in the early stages of divergence between Mexican ducks and mallards. Mol. Ecol. 2015, 24, 5364–5378. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.L.; Lavretsky, P.; DaCosta, J.M.; Bielefeld, R.R.; Feddersen, J.C.; Sorenson, M.D. Population genomic data delineate conservation units in mottled ducks (Anas fulvigula). Biol. Conserv. 2016, 203, 272–281. [Google Scholar] [CrossRef]
- Lancaster, J.D.; Anderson, T.; Brasher, M.G.; Conway, W.C.; DeMaso, S.J.; Moon, J.A.; Ringelman, K.M.; Wilson, B.C. Gulf Coast Joint Venture Mottled Duck Conservation Plan Update; Gulf Coast Joint Venture: Lafayette, LA, USA, 2023; p. 75. [Google Scholar]
- Brown, J.I.; Hernández, F.; Engilis, A.; Hernández-Baños, B.E.; Collins, D.; Lavretsky, P. Genomic and morphological data shed light on the complexities of shared ancestry between closely related duck species. Sci. Rep. 2022, 12, 10212. [Google Scholar] [CrossRef]
- Stutzenbaker, C.D. The Mottled Duck, Its Life History, Ecology and Management; Texas Parks and Wildlife Department: Austin, TX, USA, 1988.
- Lockwood, M.W.; Freeman, B. The TOS Handbook of Texas Birds; Texas A&M University Press: College Station, TX, USA, 2014. [Google Scholar]
- Coxen, C.L.; Frey, J.K.; Carleton, S.A.; Collins, D.P. Species distribution models for a migratory bird based on citizen science and satellite tracking data. Glob. Ecol. Conserv. 2017, 11, 298–311. [Google Scholar] [CrossRef]
- Peters, J.L.; Sonsthagen, S.A.; Lavretsky, P.; Rezsutek, M.; Johnson, W.P.; McCracken, K.G. Interspecific hybridization contributes to high genetic diversity and apparent effective population size in an endemic population of mottled ducks (Anas fulvigula maculosa). Conserv. Genet. 2014, 15, 509–520. [Google Scholar] [CrossRef]
- Baldassarre, G. Ducks, Geese, and Swans of North America; Johns Hopkins University Press: Baltimore, MD, USA, 2014. [Google Scholar]
- Sorenson, M.D.; Fleischer, R.C. Multiple independent transpositions of mitochondrial DNA control region sequences to the nucleus. Proc. Natl. Acad. Sci. USA 1996, 93, 15239–15243. [Google Scholar] [CrossRef] [PubMed]
- Sorenson, M.D.; Ast, J.C.; Dimcheff, D.E.; Yuri, T.; Mindell, D.P. Primers for a PCR-based approach to mitochondrial genome sequencing in birds and other vertebrates. Mol. Phylogenetics Evol. 1999, 12, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D. Popart: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Lavretsky, P.; Hernandez, F.; Swale, T.; Mohl, J.E. Chromosomal-level reference genome of a wild North American mallard. G3 Genes Genomes Genet. 2023, 13, jkad171. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Inference of human population history from individual whole-genome sequences. Nature 2011, 475, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Admixture 1.22 Software Manual. 2012. Available online: https://www.yumpu.com/en/document/view/21419722/admixture-122-software-manual-ucla-human-genetics (accessed on 11 April 2024).
- Alexander, D.H.; Lange, K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinform. 2011, 12, 246. [Google Scholar] [CrossRef]
- Zhou, H.; Alexander, D.; Lange, K. A quasi-Newton acceleration for high-dimensional optimization algorithms. Stat. Comput. 2011, 21, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Malinsky, M.; Trucchi, E.; Lawson, D.J.; Falush, D. RADpainter and fineRADstructure: Population inference from RADseq data. Mol. Biol. Evol. 2018, 35, 1284–1290. [Google Scholar] [CrossRef] [PubMed]
- Malinsky, M.; Svardal, H.; Tyers, A.M.; Miska, E.A.; Genner, M.J.; Turner, G.F.; Durbin, R. Whole-genome sequences of Malawi cichlids reveal multiple radiations interconnected by gene flow. Nat. Ecol. Evol. 2018, 2, 1940–1955. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.O.I. The Mexican Duck in Mexico: Natural History, Distribution, and Population Status; Colorado State University: Fort Collins, CO, USA, 1980. [Google Scholar]
- Moon, J.; Lehnen, S.; Metzger, K.; Squires, M.; Brasher, M.; Wilson, B.; Conway, W.; Haukos, D.; Davis, B.; Rohwer, F.; et al. Projected impact of sea-level rise and urbanization on mottled duck (Anas fulvigula) habitat along the Gulf Coast of Louisiana and Texas through 2100. Ecol. Indic. 2021, 132, 108276. [Google Scholar] [CrossRef]
- Enwright, N.M.; Cheney, W.C.; Evans, K.O.; Thurman, H.R.; Woodrey, M.S.; Fournier, A.M.V.; Moon, J.A.; Levy, H.E.; Cox, J.A.; Kappes, P.J.; et al. Mapping high marsh and salt pannes/flats along the northern Gulf of Mexico coast. Geocarto Int. 2023, 38, 2285354. [Google Scholar] [CrossRef]
- Ross, B.E.; Haukos, D.A.; Walther, P. Quantifying changes and influences on mottled duck density in Texas. J. Wildl. Manag. 2018, 82, 374–382. [Google Scholar] [CrossRef]
- Weiss-Lehman, C.; Shaw, A.K. Spatial population structure determines extinction risk in climate-induced range shifts. Am. Nat. 2020, 195, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.J. Sea-level rise implications for coastal regions. J. Coast. Res. 2013, 63, 184–196. [Google Scholar] [CrossRef]
- USFWS. Adaptive Harvest Management: 2023 Hunting Season; U.S. Department of the Interior: Washington, DC, USA, 2022; p. 68.
- Pérez-Arteaga, A.; Gaston, K. Wildfowl population trends in Mexico, 1961–2000: A basis for conservation planning. Biol. Conserv. 2004, 115, 343–355. [Google Scholar] [CrossRef]
- Perez-Arteaga, A.; Gaston, K.J.; Kershaw, M. Population trends and priority conservation sites for Mexican Duck Anas diazi. Bird Conserv. Int. 2002, 12, 35–52. [Google Scholar] [CrossRef]
- Turbek, S.P.; Taylor, S.A. Hybridization provides climate resilience. Nat. Clim. Chang. 2023, 13, 212–213. [Google Scholar] [CrossRef]
- Lavretsky, P. Population Genomics Provides Key Insights into Admixture, Speciation, and Evolution of Closely Related Ducks of the Mallard Complex. In Population Genomics: Wildlife, Population Genomics; Hohenlohe, P., Rajora, O.P., Eds.; Springer: Cham, Switzerland, 2020. [Google Scholar]
- Champagnon, J.; Elmberg, J.; Guillemain, M.; Lavretsky, P.; Clark, R.G.; Söderquist, P. Silent domestication of wildlife in the Anthropocene: The mallard as a case study. Biol. Conserv. 2023, 288, 110354. [Google Scholar] [CrossRef]
- Lavretsky, P.; Duenez, E.; Kneece, M.; Kaminski, R.M. Population Genetics of a Translocated Population of Mottled Ducks and Allies. J. Wildl. Manag. 2021, 85, 1616–1627. [Google Scholar] [CrossRef]
- Cooper, T.; Fronczak, D. Central Flyway Harvest and Population Survey Data Book 2022. 2023. Available online: https://www.fws.gov/sites/default/files/documents/central_flyway_databook_2022.pdf (accessed on 11 April 2024).
- Ford, R.J.; Selman, W.; Taylor, S.S. Hybridization between Mottled Ducks (Anas fulvigula maculosa) and Mallards (A. platyrhynchos) in the western Gulf Coast region. Condor 2017, 119, 683–696. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lavretsky, P.; Kraai, K.J.; Butler, D.; Morel, J.; VonBank, J.A.; Marty, J.R.; Musni, V.M.; Collins, D.P. Human-Induced Range Expansions Result in a Recent Hybrid Zone between Sister Species of Ducks. Genes 2024, 15, 651. https://doi.org/10.3390/genes15060651
Lavretsky P, Kraai KJ, Butler D, Morel J, VonBank JA, Marty JR, Musni VM, Collins DP. Human-Induced Range Expansions Result in a Recent Hybrid Zone between Sister Species of Ducks. Genes. 2024; 15(6):651. https://doi.org/10.3390/genes15060651
Chicago/Turabian StyleLavretsky, Philip, Kevin J. Kraai, David Butler, James Morel, Jay A. VonBank, Joseph R. Marty, Vergie M. Musni, and Daniel P. Collins. 2024. "Human-Induced Range Expansions Result in a Recent Hybrid Zone between Sister Species of Ducks" Genes 15, no. 6: 651. https://doi.org/10.3390/genes15060651





