The Activity of YCA1 Metacaspase Is Regulated by Reactive Sulfane Sulfur via Persulfidation in Saccharomyces cerevisiae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Materials
2.2. S. cerevisiae BY4742 Mutant Construction
2.3. Protein Expression and Purification
2.4. Analysis of Inhibitory Effect of RSS on YCA1 Cleavage Activity
2.5. Protein LC-MS/MS Analysis
2.6. Analysis of Growth Curve
2.7. Analysis of Intracellular RSS Level
2.8. Analysis of YCA1 Activity In Vivo
2.9. Chronological Lifespan Assay
2.10. CLS-Driven Apoptosis Assay
2.11. Transcriptomic Analysis
3. Results
3.1. RSS Inhibited Proteolytic Activity of YCA1 In Vitro
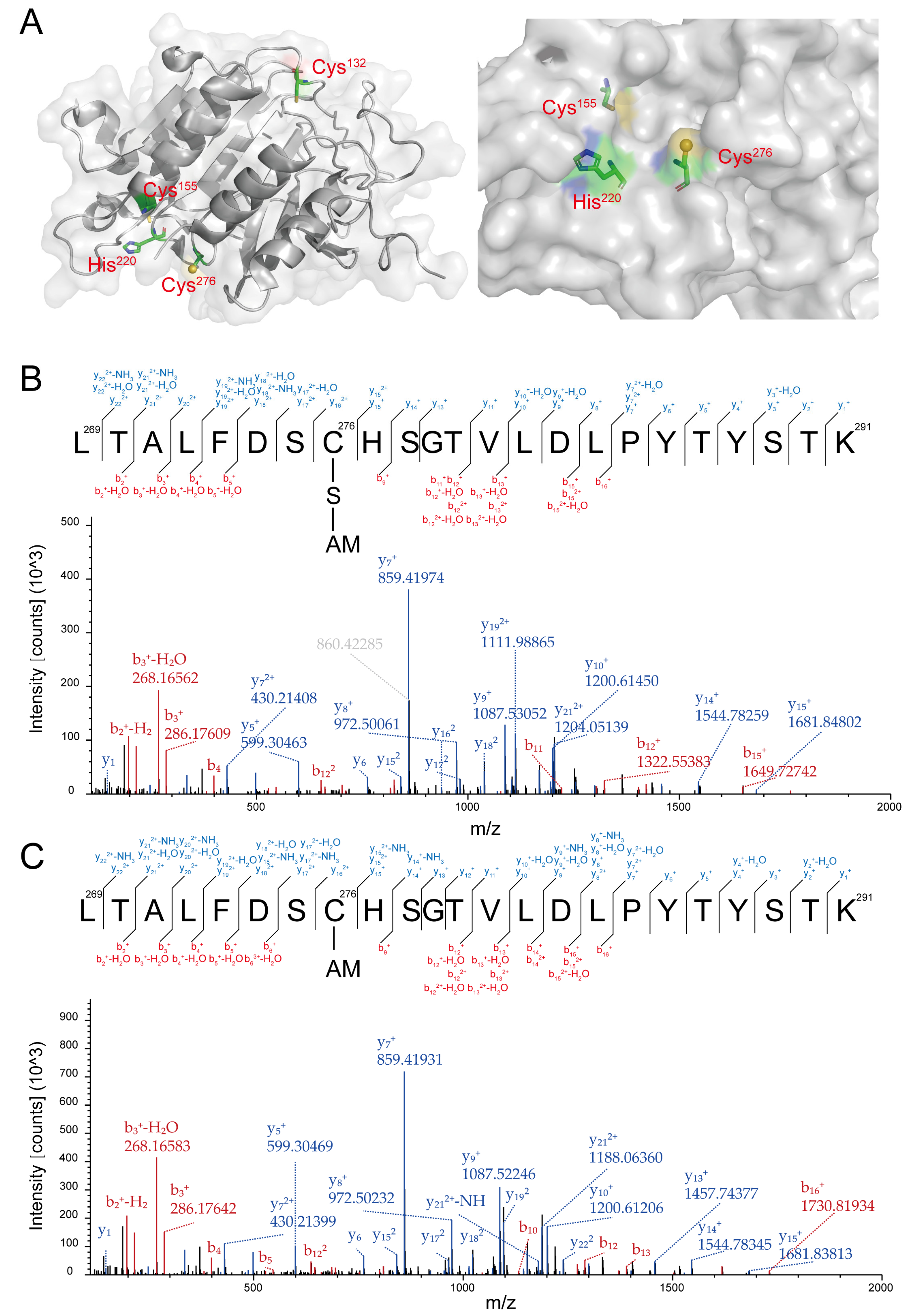
3.2. Cys276 of YCA1 Was Reversibly Modified by RSS
3.3. RSS Inhibited Proteolytic Activity of YCA1 In Vivo
3.4. Increased Proteolytic Activity of YCA1 Led to Short CLS and Early Apoptosis
3.5. Systematic Analysis of Gene Expression Changes in YCA1-Activated Strain
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, Z.P.; Ji, Q.Z.; Ren, J.; Yan, P.Z.; Wu, Z.M.; Wang, S.; Sun, L.; Wang, Z.H.; Li, J.M.; Sun, G.Q.; et al. Large-scale chromatin reorganization reactivates placenta-specific genes that drive cellular aging. Dev. Cell 2022, 57, 1347–1368.e12. [Google Scholar] [CrossRef]
- Zhang, L.; Pitcher, L.E.; Yousefzadeh, M.J.; Niedernhofer, L.J.; Robbins, P.D.; Zhu, Y. Cellular senescence: A key therapeutic target in aging and diseases. J. Clin. Investig. 2022, 132, e158450. [Google Scholar] [CrossRef] [PubMed]
- Murphy, B.M.; Burd, C.E. The aging lung microenvironment awakens melanoma metastases. Cancer Cell 2022, 40, 815–817. [Google Scholar] [CrossRef]
- Priault, M.; Camougrand, N.; Kinnally, K.W.; Vallette, F.M.; Manon, S. Yeast as a tool to study Bax/mitochondrial interactions in cell death. FEMS Yeast Res. 2003, 4, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Kaya, A.; Lobanov, A.V.; Gladyshev, V.N. Evidence that mutation accumulation does not cause aging in Saccharomyces cerevisiae. Aging Cell 2015, 14, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Büttner, S.; Habernig, L.; Broeskamp, F.; Ruli, D.; Vögtle, F.N.; Vlachos, M.; Macchi, F.; Küttner, V.; Carmona-Gutierrez, D.; Eisenberg, T.; et al. Endonuclease G mediates α-synuclein cytotoxicity during Parkinson’s disease. EMBO J. 2013, 32, 3041–3054. [Google Scholar] [CrossRef] [PubMed]
- Sudharshan, S.J.; Narayanan, A.K.; Princilly, J.; Dyavaiah, M.; Nagegowda, D.A. Betulinic acid mitigates oxidative stress-mediated apoptosis and enhances longevity in the yeast Saccharomyces cerevisiae model. Free Radic. Res. 2022, 56, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Carmona-Gutiérrez, D.; Bauer, M.A.; Ring, J.; Knauer, H.; Eisenberg, T.; Büttner, S.; Ruckenstuhl, C.; Reisenbichler, A.; Magnes, C.; Rechberger, G.N.; et al. The propeptide of yeast cathepsin D inhibits programmed necrosis. Cell Death Dis. 2011, 2, e161. [Google Scholar] [CrossRef] [PubMed]
- Sudharshan, S.J.; Veerabhadrappa, B.; Subramaniyan, S.; Dyavaiah, M. Astaxanthin enhances the longevity of Saccharomyces cerevisiae by decreasing oxidative stress and apoptosis. FEMS Yeast Res. 2019, 19, foy113. [Google Scholar] [CrossRef]
- Lam, D.K.; Sherlock, G. Yca1 metacaspase: Diverse functions determine how yeast live and let die. FEMS Yeast Res. 2023, 23, foad022. [Google Scholar] [CrossRef]
- Lin, S.J.; Austriaco, N. Aging and cell death in the other yeasts, Schizosaccharomyces pombe and Candida albicans. FEMS Yeast Res. 2014, 14, 119–135. [Google Scholar] [CrossRef] [PubMed]
- Madeo, F.; Herker, E.; Maldener, C.; Wissing, S.; Lächelt, S.; Herian, M.; Fehr, M.; Lauber, K.; Sigrist, S.J.; Wesselborg, S.; et al. A caspase-related protease regulates apoptosis in yeast. Mol. Cell 2002, 9, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Hill, S.M.; Hao, X.X.; Liu, B.D.; Nyström, T. Life-span extension by a metacaspase in the yeast Saccharomyces cerevisiae. Science 2014, 344, 1389–1392. [Google Scholar] [CrossRef] [PubMed]
- Lau, N.; Pluth, M.D. Reactive sulfur species (RSS): Persulfides, polysulfides, potential, and problems. Curr. Opin. Chem. Biol. 2019, 49, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; Yu, R.H.; Wu, L.Y.; Yang, G.D. Hydrogen sulfide signaling in regulation of cell behaviors. Nitric Oxide-Biol. Chem. 2020, 103, 9–19. [Google Scholar] [CrossRef]
- Ran, M.X.; Wang, T.Q.; Shao, M.; Chen, Z.G.; Liu, H.W.; Xia, Y.Z.; Xun, L.Y. Sensitive method for reliable quantification of sulfane sulfur in biological samples. Anal. Chem. 2019, 91, 11981–11986. [Google Scholar] [CrossRef]
- Wang, Q.D.; Chen, Z.G.; Zhang, X.; Xin, Y.P.; Xia, Y.Z.; Xun, L.Y.; Liu, H.W. Rhodanese Rdl2 produces reactive sulfur species to protect mitochondria from reactive oxygen species. Free Radic. Biol. Med. 2021, 177, 287–298. [Google Scholar] [CrossRef]
- Akaike, T.; Ida, T.; Wei, F.Y.; Nishida, M.; Kumagai, Y.; Alam, M.M.; Ihara, H.; Sawa, T.; Matsunaga, T.; Kasamatsu, S.; et al. Cysteinyl-tRNA synthetase governs cysteine polysulfidation and mitochondrial bioenergetics. Nat. Commun. 2017, 8, 1177. [Google Scholar] [CrossRef]
- Yadav, P.K.; Martinov, M.; Vitvitsky, V.; Seravalli, J.; Wedmann, R.; Filipovic, M.R.; Banerjee, R. Biosynthesis and reactivity of Cysteine persulfides in signaling. J. Am. Chem. Soc. 2016, 138, 289–299. [Google Scholar] [CrossRef]
- Shah, A.A.; Liu, B.H.; Tang, Z.H.; Wang, W.; Yang, W.J.; Hu, Q.J.; Liu, Y.; Zhang, N.H.; Liu, K. Hydrogen sulfide treatment at the late growth stage of Saccharomyces cerevisiae extends chronological lifespan. Aging 2021, 13, 9859–9873. [Google Scholar] [CrossRef]
- Hine, C.; Harputlugil, E.; Zhang, Y.; Ruckenstuhl, C.; Lee, B.C.; Brace, L.; Longchamp, A.; Treviño-Villarreal, J.H.; Mejia, P.; Ozaki, C.K.; et al. Endogenous hydrogen sulfide production is essential for dietary restriction benefits. Cell 2015, 160, 132–144. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, A.K.; Gadalla, M.M.; Sen, N.; Kim, S.; Mu, W.; Gazi, S.K.; Barrow, R.K.; Yang, G.; Wang, R.; Snyder, S.H. H2S signals through protein S-sulfhydration. Sci. Signal. 2009, 2, ra72. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.C.; Walsh, B.J.C.; Flores-Mireles, A.L.; Peng, H.; Zhang, Y.F.; Zhang, Y.X.; Trinidad, J.C.; Hultgren, S.J.; Giedroc, D.P. Hydrogen sulfide sensing through reactive sulfur species (RSS) and nitroxyl (HNO) in Enterococcus faecalis. ACS Chem. Biol. 2018, 13, 1610–1620. [Google Scholar] [CrossRef] [PubMed]
- Ju, Y.; Untereiner, A.; Wu, L.; Yang, G. H2S-induced S-sulfhydration of pyruvate carboxylase contributes to gluconeogenesis in liver cells. Biochim. Biophys. Acta Gen. Subj. 2015, 1850, 2293–2303. [Google Scholar] [CrossRef] [PubMed]
- Untereiner, A.A.; Oláh, G.; Módis, K.; Hellmich, M.R.; Szabo, C. H2S-induced S-sulfhydration of lactate dehydrogenase a (LDHA) stimulates cellular bioenergetics in HCT116 colon cancer cells. Biochem. Pharmacol. 2017, 136, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Braunstein, I.; Engelman, R.; Yitzhaki, O.; Ziv, T.; Galardon, E.; Benhar, M. Opposing effects of polysulfides and thioredoxin on apoptosis through caspase persulfidation. J. Biol. Chem. 2020, 295, 3590–3600. [Google Scholar] [CrossRef] [PubMed]
- Luebke, J.L.; Shen, J.C.; Bruce, K.E.; Kehl-Fie, T.E.; Peng, H.; Skaar, E.P.; Giedroc, D.P. The CsoR-like sulfurtransferase repressor (CstR) is a persulfide sensor in Staphylococcus aureus. Mol. Microbiol. 2014, 94, 1343–1360. [Google Scholar] [CrossRef] [PubMed]
- Guldener, U.; Heck, S.; Fiedler, T.; Beinhauer, J.; Hegemann, J.H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996, 24, 2519–2524. [Google Scholar] [CrossRef]
- Gietz, R.D.; Woods, R.A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. In Guide to Yeast Genetics and Molecular and Cell Biology; Academic Press: Cambridge, MA, USA, 2002; Volume 350, pp. 87–96. [Google Scholar] [CrossRef]
- Xia, Y.Z.; Li, K.; Li, J.J.; Wang, T.Q.; Gu, L.C.; Xun, L.Y. T5 exonuclease-dependent assembly offers a low-cost method for efficient cloning and site-directed mutagenesis. Nucleic Acids Res. 2019, 47, e15. [Google Scholar] [CrossRef]
- Li, H.J.; Li, J.; Lü, C.J.; Xia, Y.Z.; Xin, Y.F.; Liu, H.L.; Xun, L.Y.; Liu, H.W. FisR activates σ-dependent transcription of sulfide-oxidizing genes in Cupriavidus pinatubonensis JMP134. Mol. Microbiol. 2017, 105, 373–384. [Google Scholar] [CrossRef]
- Yu, Q.L.; Ran, M.X.; Yang, Y.Q.; Liu, H.W.; Xun, L.Y.; Xia, Y.Z. Optimization of a method for detecting intracellular sulfane sulfur Levels and evaluation of reagents that affect the levels in Escherichia coli. Antioxidants 2022, 11, 1292. [Google Scholar] [CrossRef] [PubMed]
- Jung, P.P.; Zhang, Z.; Paczia, N.; Jaeger, C.; Ignac, T.; May, P.; Linster, C.L. Natural variation of chronological aging in the Saccharomyces cerevisiae species reveals diet-dependent mechanisms of life span control. npj Aging Mech. Dis. 2018, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.H.H.; Yan, C.Y.; Shi, Y.G. Crystal Structure of the Yeast Metacaspase Yca1. J. Biol. Chem. 2012, 287, 29251–29259. [Google Scholar] [CrossRef] [PubMed]
- Herker, E.; Jungwirth, H.; Lehmann, K.A.; Maldener, C.; Fröhlich, K.U.; Wissing, S.; Büttner, S.; Fehr, M.; Sigrist, S.; Madeo, F. Chronological aging leads to apoptosis in yeast. J. Cell Biol. 2004, 164, 501–507. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Zhang, X.; Du, Z.; Liu, H.; Xia, Y.; Xun, L.; Liu, H. The Activity of YCA1 Metacaspase Is Regulated by Reactive Sulfane Sulfur via Persulfidation in Saccharomyces cerevisiae. Antioxidants 2024, 13, 589. https://doi.org/10.3390/antiox13050589
Wang Q, Zhang X, Du Z, Liu H, Xia Y, Xun L, Liu H. The Activity of YCA1 Metacaspase Is Regulated by Reactive Sulfane Sulfur via Persulfidation in Saccharomyces cerevisiae. Antioxidants. 2024; 13(5):589. https://doi.org/10.3390/antiox13050589
Chicago/Turabian StyleWang, Qingda, Xiaokun Zhang, Zhuang Du, Honglei Liu, Yongzhen Xia, Luying Xun, and Huaiwei Liu. 2024. "The Activity of YCA1 Metacaspase Is Regulated by Reactive Sulfane Sulfur via Persulfidation in Saccharomyces cerevisiae" Antioxidants 13, no. 5: 589. https://doi.org/10.3390/antiox13050589




