Presenilin Deficiency Results in Cellular Cholesterol Accumulation by Impairment of Protein Glycosylation and NPC1 Function
Abstract
:1. Introduction
2. Results
2.1. Deletion of PS1 or PS2 Induces Cellular Cholesterol Accumulation
2.2. Reduced Intracellular Transport and Increased Biosynthesis of Cholesterol in PS1-KO and PS2-KO MEFs
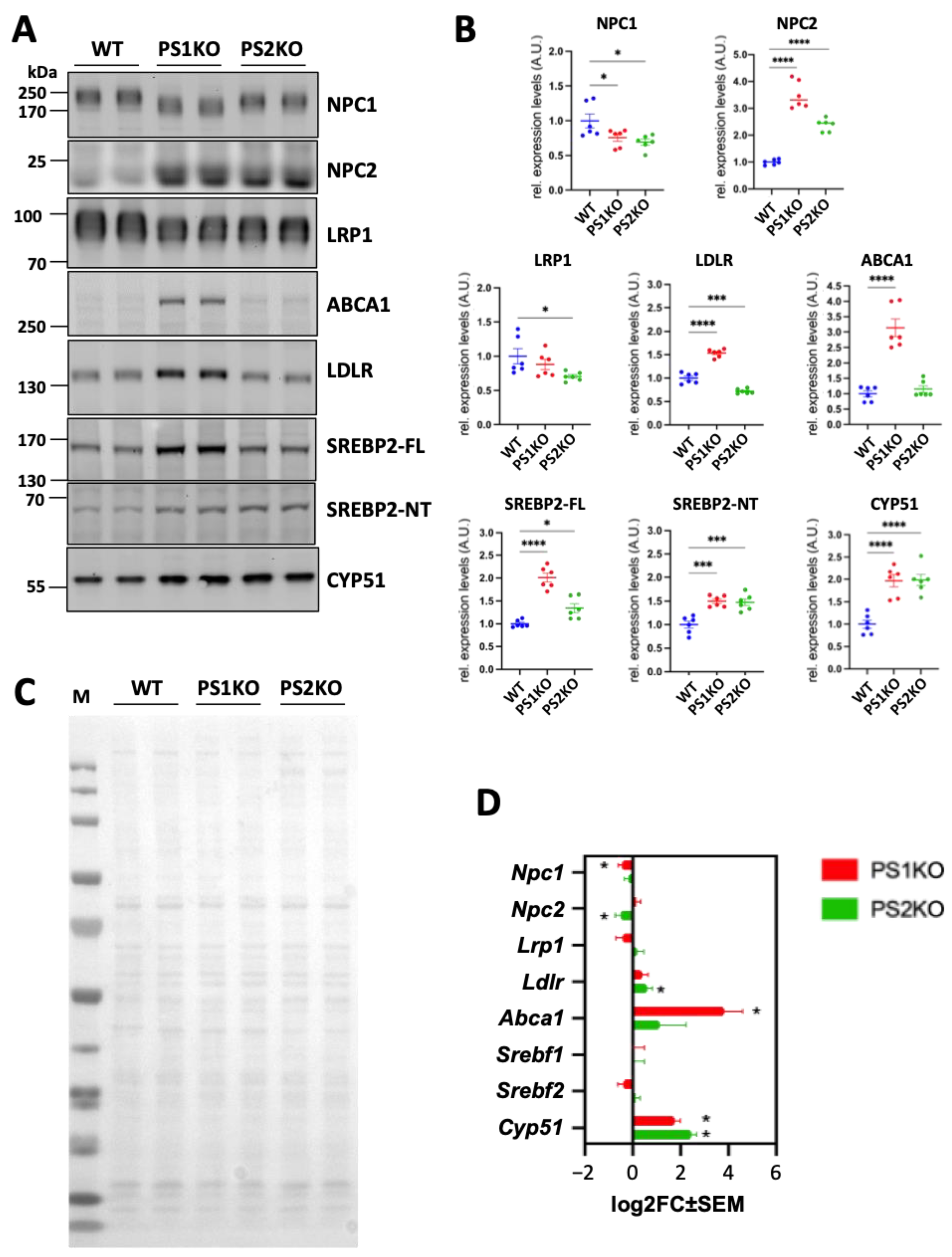
2.3. Lack of PS1 or PS2 Affects Protein Glycosylation
2.4. Impairment of Protein Glycosylation Induces Cholesterol Accumulation
2.5. Chaperone Induction Increases NPC1 Expression and Normalizes Endolysosomal Cholesterol Distribution in PS1-KO and PS2-KO MEFs
2.6. Overexpression of NPC1 Rescues Intracellular Cholesterol Accumulation in PS1-KO and PS2-KO MEFs
2.7. Effect of γ-Secretase Inhibition on NPC1 Expression
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. Cell Culture
4.3. Sample Preparation, SDS-PAGE, and Western Blotting
4.4. Immunofluorescence Staining
4.5. Imaging and Data Analysis
4.6. Sterol Mass Spectrometry Analysis
4.7. N-Glycan Structural Analysis by Mass Spectrometry
4.8. Deglycosylation of Membrane Proteins
4.9. 3′-mRNA Sequencing
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jurisch-Yaksi, N.; Sannerud, R.; Annaert, W. A Fast Growing Spectrum of Biological Functions of γ-Secretase in Development and Disease. Biochim. Biophys. Acta 2013, 1828, 2815–2827. [Google Scholar] [CrossRef] [PubMed]
- Duggan, S.P.; McCarthy, J.V. Beyond γ-Secretase Activity: The Multifunctional Nature of Presenilins in Cell Signalling Pathways. Cell Signal. 2016, 28, 1–11. [Google Scholar] [CrossRef]
- Oikawa, N.; Walter, J. Presenilins and γ-Secretase in Membrane Proteostasis. Cells 2019, 8, 209. [Google Scholar] [CrossRef]
- De Strooper, B.; Saftig, P.; Craessaerts, K.; Vanderstichele, H.; Guhde, G.; Annaert, W.; Von Figura, K.; Van Leuven, F. Deficiency of Presenilin-1 Inhibits the Normal Cleavage of Amyloid Precursor Protein. Nature 1998, 391, 387–390. [Google Scholar] [CrossRef] [PubMed]
- De Strooper, B.; Annaert, W.; Cupers, P.; Saftig, P.; Craessaerts, K.; Mumm, J.S.; Schroeter, E.H.; Schrijvers, V.; Wolfe, M.S.; Ray, W.J.; et al. A Presenilin-1-Dependent Gamma-Secretase-like Protease Mediates Release of Notch Intracellular Domain. Nature 1999, 398, 518–522. [Google Scholar] [CrossRef] [PubMed]
- De Strooper, B.; Iwatsubo, T.; Wolfe, M.S. Presenilins and γ-Secretase: Structure, Function, and Role in Alzheimer Disease. Cold Spring Harb. Perspect. Med. 2012, 2, a006304. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, M.S.; Xia, W.; Ostaszewski, B.L.; Diehl, T.S.; Kimberly, W.T.; Selkoe, D.J. Two Transmembrane Aspartates in Presenilin-1 Required for Presenilin Endoproteolysis and Gamma-Secretase Activity. Nature 1999, 398, 513–517. [Google Scholar] [CrossRef]
- Wolfe, M.S. Unraveling the Complexity of γ-Secretase. Semin. Cell Dev. Biol. 2020, 105, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Dickson, D.W. The Pathogenesis of Senile Plaques. J. Neuropathol. Exp. Neurol. 1997, 56, 321–339. [Google Scholar] [CrossRef] [PubMed]
- Levy-Lahad, E.; Wasco, W.; Poorkaj, P.; Romano, D.M.; Oshima, J.; Pettingell, W.H.; Yu, C.E.; Jondro, P.D.; Schmidt, S.D.; Wang, K. Candidate Gene for the Chromosome 1 Familial Alzheimer’s Disease Locus. Science 1995, 269, 973–977. [Google Scholar] [CrossRef] [PubMed]
- Rogaev, E.I.; Sherrington, R.; Rogaeva, E.A.; Levesque, G.; Ikeda, M.; Liang, Y.; Chi, H.; Lin, C.; Holman, K.; Tsuda, T. Familial Alzheimer’s Disease in Kindreds with Missense Mutations in a Gene on Chromosome 1 Related to the Alzheimer’s Disease Type 3 Gene. Nature 1995, 376, 775–778. [Google Scholar] [CrossRef] [PubMed]
- Sherrington, R.; Rogaev, E.I.; Liang, Y.; Rogaeva, E.A.; Levesque, G.; Ikeda, M.; Chi, H.; Lin, C.; Li, G.; Holman, K.; et al. Cloning of a Gene Bearing Missense Mutations in Early-Onset Familial Alzheimer’s Disease. Nature 1995, 375, 754–760. [Google Scholar] [CrossRef] [PubMed]
- De Strooper, B. Loss-of-Function Presenilin Mutations in Alzheimer Disease. Talking Point on the Role of Presenilin Mutations in Alzheimer Disease. EMBO Rep. 2007, 8, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Kelleher, R.J. The Presenilin Hypothesis of Alzheimer’s Disease: Evidence for a Loss-of-Function Pathogenic Mechanism. Proc. Natl. Acad. Sci. USA 2007, 104, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Heilig, E.A.; Xia, W.; Shen, J.; Kelleher, R.J. A Presenilin-1 Mutation Identified in Familial Alzheimer Disease with Cotton Wool Plaques Causes a Nearly Complete Loss of Gamma-Secretase Activity. J. Biol. Chem. 2010, 285, 22350–22359. [Google Scholar] [CrossRef] [PubMed]
- Xia, D.; Watanabe, H.; Wu, B.; Lee, S.H.; Li, Y.; Tsvetkov, E.; Bolshakov, V.Y.; Shen, J.; Kelleher, R.J. Presenilin-1 Knockin Mice Reveal Loss-of-Function Mechanism for Familial Alzheimer’s Disease. Neuron 2015, 85, 967–981. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhou, R.; Yang, G.; Shi, Y. Analysis of 138 Pathogenic Mutations in Presenilin-1 on the in Vitro Production of Aβ42 and Aβ40 Peptides by γ-Secretase. Proc. Natl. Acad. Sci. USA 2017, 114, E476–E485. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, M.S. Dysfunctional γ-Secretase in Familial Alzheimer’s Disease. Neurochem. Res. 2019, 44, 5–11. [Google Scholar] [CrossRef]
- Hata, S.; Fujishige, S.; Araki, Y.; Taniguchi, M.; Urakami, K.; Peskind, E.; Akatsu, H.; Araseki, M.; Yamamoto, K.; Martins, R.N.; et al. Alternative Processing of γ-Secretase Substrates in Common Forms of Mild Cognitive Impairment and Alzheimer’s Disease: Evidence for γ-Secretase Dysfunction. Ann. Neurol. 2011, 69, 1026–1031. [Google Scholar] [CrossRef] [PubMed]
- Kakuda, N.; Yamaguchi, H.; Akazawa, K.; Hata, S.; Suzuki, T.; Hatsuta, H.; Murayama, S.; Funamoto, S.; Ihara, Y. γ-Secretase Activity Is Associated with Braak Senile Plaque Stages. Am. J. Pathol. 2020, 190, 1323–1331. [Google Scholar] [CrossRef] [PubMed]
- Quintero-Monzon, O.; Martin, M.M.; Fernandez, M.A.; Cappello, C.A.; Krzysiak, A.J.; Osenkowski, P.; Wolfe, M.S. Dissociation between the Processivity and Total Activity of γ-Secretase: Implications for the Mechanism of Alzheimer’s Disease-Causing Presenilin Mutations. Biochemistry 2011, 50, 9023–9035. [Google Scholar] [CrossRef] [PubMed]
- Szaruga, M.; Munteanu, B.; Lismont, S.; Veugelen, S.; Horré, K.; Mercken, M.; Saido, T.C.; Ryan, N.S.; De Vos, T.; Savvides, S.N.; et al. Alzheimer’s-Causing Mutations Shift Aβ Length by Destabilizing γ-Secretase-Aβn Interactions. Cell 2017, 170, 443–456.e14. [Google Scholar] [CrossRef] [PubMed]
- Chávez-Gutiérrez, L.; Szaruga, M. Mechanisms of Neurodegeneration—Insights from Familial Alzheimer’s Disease. Semin. Cell Dev. Biol. 2020, 105, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Saura, C.A.; Choi, S.-Y.; Beglopoulos, V.; Malkani, S.; Zhang, D.; Shankaranarayana Rao, B.S.; Chattarji, S.; Kelleher, R.J.; Kandel, E.R.; Duff, K.; et al. Loss of Presenilin Function Causes Impairments of Memory and Synaptic Plasticity Followed by Age-Dependent Neurodegeneration. Neuron 2004, 42, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Tabuchi, K.; Chen, G.; Südhof, T.C.; Shen, J. Conditional Forebrain Inactivation of Nicastrin Causes Progressive Memory Impairment and Age-Related Neurodegeneration. J. Neurosci. 2009, 29, 7290–7301. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.; Iqbal, M.; Zheng, J.; Wines-Samuelson, M.; Shen, J. Partial Loss of Presenilin Impairs Age-Dependent Neuronal Survival in the Cerebral Cortex. J. Neurosci. 2014, 34, 15912–15922. [Google Scholar] [CrossRef]
- Acx, H.; Serneels, L.; Radaelli, E.; Muyldermans, S.; Vincke, C.; Pepermans, E.; Müller, U.; Chávez-Gutiérrez, L.; De Strooper, B. Inactivation of γ-Secretases Leads to Accumulation of Substrates and Non-Alzheimer Neurodegeneration. EMBO Mol. Med. 2017, 9, 1088–1099. [Google Scholar] [CrossRef] [PubMed]
- Grimm, M.O.W.; Grimm, H.S.; Pätzold, A.J.; Zinser, E.G.; Halonen, R.; Duering, M.; Tschäpe, J.A.; De Strooper, B.; Müller, U.; Shen, J.; et al. Regulation of Cholesterol and Sphingomyelin Metabolism by Amyloid-Beta and Presenilin. Nat. Cell Biol. 2005, 7, 1118–1123. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zerbinatti, C.V.; Zhang, J.; Hoe, H.-S.; Wang, B.; Cole, S.L.; Herz, J.; Muglia, L.; Bu, G. Amyloid Precursor Protein Regulates Brain Apolipoprotein E and Cholesterol Metabolism through Lipoprotein Receptor LRP1. Neuron 2007, 56, 66–78. [Google Scholar] [CrossRef]
- Tamboli, I.Y.; Prager, K.; Thal, D.R.; Thelen, K.M.; Dewachter, I.; Pietrzik, C.U.; St George-Hyslop, P.; Sisodia, S.S.; De Strooper, B.; Heneka, M.T.; et al. Loss of Gamma-Secretase Function Impairs Endocytosis of Lipoprotein Particles and Membrane Cholesterol Homeostasis. J. Neurosci. 2008, 28, 12097–12106. [Google Scholar] [CrossRef] [PubMed]
- Area-Gomez, E.; Del Carmen Lara Castillo, M.; Tambini, M.D.; Guardia-Laguarta, C.; de Groof, A.J.C.; Madra, M.; Ikenouchi, J.; Umeda, M.; Bird, T.D.; Sturley, S.L.; et al. Upregulated Function of Mitochondria-Associated ER Membranes in Alzheimer Disease. EMBO J. 2012, 31, 4106–4123. [Google Scholar] [CrossRef]
- Gutierrez, E.; Lütjohann, D.; Kerksiek, A.; Fabiano, M.; Oikawa, N.; Kuerschner, L.; Thiele, C.; Walter, J. Importance of γ-Secretase in the Regulation of Liver X Receptor and Cellular Lipid Metabolism. Life Sci. Alliance 2020, 3, e201900521. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.N.; Son, D.J.; Lee, J.W.; Hwang, D.Y.; Kim, Y.K.; Cho, J.S.; Lee, U.S.; Yoo, H.S.; Moon, D.C.; Oh, K.W.; et al. Mutant Presenilin 2 Causes Abnormality in the Brain Lipid Profile in the Development of Alzheimer’s Disease. Arch. Pharm. Res. 2006, 29, 884–889. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.Y.; Kwon, O.-H.; Park, M.K.; Kim, T.-W.; Chung, S. Elevated Cellular Cholesterol in Familial Alzheimer’s Presenilin 1 Mutation Is Associated with Lipid Raft Localization of β-Amyloid Precursor Protein. PLoS ONE 2019, 14, e0210535. [Google Scholar] [CrossRef] [PubMed]
- Churchward, M.A.; Coorssen, J.R. Cholesterol, Regulated Exocytosis and the Physiological Fusion Machine. Biochem. J. 2009, 423, 1–14. [Google Scholar] [CrossRef]
- Simons, K.; Gerl, M.J. Revitalizing Membrane Rafts: New Tools and Insights. Nat. Rev. Mol. Cell Biol. 2010, 11, 688–699. [Google Scholar] [CrossRef] [PubMed]
- Enrich, C.; Rentero, C.; Hierro, A.; Grewal, T. Role of Cholesterol in SNARE-Mediated Trafficking on Intracellular Membranes. J. Cell Sci. 2015, 128, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Tharkeshwar, A.K.; Trekker, J.; Vermeire, W.; Pauwels, J.; Sannerud, R.; Priestman, D.A.; Te Vruchte, D.; Vints, K.; Baatsen, P.; Decuypere, J.-P.; et al. A Novel Approach to Analyze Lysosomal Dysfunctions through Subcellular Proteomics and Lipidomics: The Case of NPC1 Deficiency. Sci. Rep. 2017, 7, 41408. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, S.; Sillence, D.J. Niemann-Pick Type C Disease: Cellular Pathology and Pharmacotherapy. J. Neurochem. 2020, 153, 674–692. [Google Scholar] [CrossRef] [PubMed]
- Auer, I.A.; Schmidt, M.L.; Lee, V.M.; Curry, B.; Suzuki, K.; Shin, R.W.; Pentchev, P.G.; Carstea, E.D.; Trojanowski, J.Q. Paired Helical Filament Tau (PHFtau) in Niemann-Pick Type C Disease Is Similar to PHFtau in Alzheimer’s Disease. Acta Neuropathol. 1995, 90, 547–551. [Google Scholar] [CrossRef]
- Love, S.; Bridges, L.R.; Case, C.P. Neurofibrillary Tangles in Niemann-Pick Disease Type C. Brain 1995, 118 Pt 1, 119–129. [Google Scholar] [CrossRef]
- Suzuki, K.; Parker, C.C.; Pentchev, P.G.; Katz, D.; Ghetti, B.; D’Agostino, A.N.; Carstea, E.D. Neurofibrillary Tangles in Niemann-Pick Disease Type C. Acta Neuropathol. 1995, 89, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Saura, C.A.; Choi, S.Y.; Sun, L.D.; Yang, X.; Handler, M.; Kawarabayashi, T.; Younkin, L.; Fedeles, B.; Wilson, M.A.; et al. APP Processing and Synaptic Plasticity in Presenilin-1 Conditional Knockout Mice. Neuron 2001, 31, 713–726. [Google Scholar] [CrossRef] [PubMed]
- Herreman, A.; Hartmann, D.; Annaert, W.; Saftig, P.; Craessaerts, K.; Serneels, L.; Umans, L.; Schrijvers, V.; Checler, F.; Vanderstichele, H.; et al. Presenilin 2 Deficiency Causes a Mild Pulmonary Phenotype and No Changes in Amyloid Precursor Protein Processing but Enhances the Embryonic Lethal Phenotype of Presenilin 1 Deficiency. Proc. Natl. Acad. Sci. USA 1999, 96, 11872–11877. [Google Scholar] [CrossRef] [PubMed]
- Kishimoto, T.; Ishitsuka, R.; Kobayashi, T. Detectors for Evaluating the Cellular Landscape of Sphingomyelin- and Cholesterol-Rich Membrane Domains. Biochim. Biophys. Acta 2016, 1861, 812–829. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Jiang, L.; Yang, H.; Song, B.-L. Routes and Mechanisms of Post-Endosomal Cholesterol Trafficking: A Story That Never Ends. Traffic 2017, 18, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Heybrock, S.; Neculai, D.; Saftig, P. Cholesterol Handling in Lysosomes and Beyond. Trends Cell Biol. 2020, 30, 452–466. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.-Y.; Chang, C.C.Y.; Ohgami, N.; Yamauchi, Y. Cholesterol Sensing, Trafficking, and Esterification. Annu. Rev. Cell Dev. Biol. 2006, 22, 129–157. [Google Scholar] [CrossRef]
- Rawson, R.B. The SREBP Pathway--Insights from Insigs and Insects. Nat. Rev. Mol. Cell Biol. 2003, 4, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Sato, R. Sterol Metabolism and SREBP Activation. Arch. Biochem. Biophys. 2010, 501, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Harrison, K.D.; Miao, R.Q.; Fernandez-Hernándo, C.; Suárez, Y.; Dávalos, A.; Sessa, W.C. Nogo-B Receptor Stabilizes Niemann-Pick Type C2 Protein and Regulates Intracellular Cholesterol Trafficking. Cell Metab. 2009, 10, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.-J.; Chen, T.-L.; Lee, T.-S.; Wang, H.-A.; Wang, C.-K.; Liao, L.-Y.; Liu, R.-S.; Huang, S.-F.; Chen, Y.-M.A. Glycine N-Methyltransferase Deficiency Affects Niemann-Pick Type C2 Protein Stability and Regulates Hepatic Cholesterol Homeostasis. Mol. Med. 2012, 18, 412–422. [Google Scholar] [CrossRef] [PubMed]
- Gelsthorpe, M.E.; Baumann, N.; Millard, E.; Gale, S.E.; Langmade, S.J.; Schaffer, J.E.; Ory, D.S. Niemann-Pick Type C1 I1061T Mutant Encodes a Functional Protein That Is Selected for Endoplasmic Reticulum-Associated Degradation Due to Protein Misfolding. J. Biol. Chem. 2008, 283, 8229–8236. [Google Scholar] [CrossRef]
- Nakasone, N.; Nakamura, Y.S.; Higaki, K.; Oumi, N.; Ohno, K.; Ninomiya, H. Endoplasmic Reticulum-Associated Degradation of Niemann-Pick C1: Evidence for the Role of Heat Shock Proteins and Identification of Lysine Residues That Accept Ubiquitin. J. Biol. Chem. 2014, 289, 19714–19725. [Google Scholar] [CrossRef] [PubMed]
- Herreman, A.; Van Gassen, G.; Bentahir, M.; Nyabi, O.; Craessaerts, K.; Mueller, U.; Annaert, W.; De Strooper, B. Gamma-Secretase Activity Requires the Presenilin-Dependent Trafficking of Nicastrin through the Golgi Apparatus but Not Its Complex Glycosylation. J. Cell Sci. 2003, 116, 1127–1136. [Google Scholar] [CrossRef] [PubMed]
- Leem, J.Y.; Vijayan, S.; Han, P.; Cai, D.; Machura, M.; Lopes, K.O.; Veselits, M.L.; Xu, H.; Thinakaran, G. Presenilin 1 Is Required for Maturation and Cell Surface Accumulation of Nicastrin. J. Biol. Chem. 2002, 277, 19236–19240. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.-S.; Tandon, A.; Chen, F.; Yu, G.; Yu, H.; Arawaka, S.; Hasegawa, H.; Duthie, M.; Schmidt, S.D.; Ramabhadran, T.V.; et al. Mature Glycosylation and Trafficking of Nicastrin Modulate Its Binding to Presenilins. J. Biol. Chem. 2002, 277, 28135–28142. [Google Scholar] [CrossRef] [PubMed]
- Tomita, T.; Katayama, R.; Takikawa, R.; Iwatsubo, T. Complex N-Glycosylated Form of Nicastrin Is Stabilized and Selectively Bound to Presenilin Fragments. FEBS Lett. 2002, 520, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Edbauer, D.; Winkler, E.; Haass, C.; Steiner, H. Presenilin and Nicastrin Regulate Each Other and Determine Amyloid Beta-Peptide Production via Complex Formation. Proc. Natl. Acad. Sci. USA 2002, 99, 8666–8671. [Google Scholar] [CrossRef] [PubMed]
- Kimberly, W.T.; LaVoie, M.J.; Ostaszewski, B.L.; Ye, W.; Wolfe, M.S.; Selkoe, D.J. Complex N-Linked Glycosylated Nicastrin Associates with Active Gamma-Secretase and Undergoes Tight Cellular Regulation. J. Biol. Chem. 2002, 277, 35113–35117. [Google Scholar] [CrossRef]
- Thimiri Govinda Raj, D.B.; Ghesquière, B.; Tharkeshwar, A.K.; Coen, K.; Derua, R.; Vanderschaeghe, D.; Rysman, E.; Bagadi, M.; Baatsen, P.; De Strooper, B.; et al. A Novel Strategy for the Comprehensive Analysis of the Biomolecular Composition of Isolated Plasma Membranes. Mol. Syst. Biol. 2011, 7, 541. [Google Scholar] [CrossRef] [PubMed]
- Stanley, P.; Moremen, K.W.; Lewis, N.E.; Taniguchi, N.; Aebi, M. N-Glycans. In Essentials of Glycobiology; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Mohnen, D., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2022; ISBN 978-1-62182-421-3. [Google Scholar]
- Vocadlo, D.J.; Lowary, T.L.; Bertozzi, C.R.; Schnaar, R.L.; Esko, J.D. Chemical Tools for Inhibiting Glycosylation. In Essentials of Glycobiology; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Mohnen, D., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2022; ISBN 978-1-62182-421-3. [Google Scholar]
- Sokol, J.; Blanchette-Mackie, J.; Kruth, H.S.; Dwyer, N.K.; Amende, L.M.; Butler, J.D.; Robinson, E.; Patel, S.; Brady, R.O.; Comly, M.E. Type C Niemann-Pick Disease. Lysosomal Accumulation and Defective Intracellular Mobilization of Low Density Lipoprotein Cholesterol. J. Biol. Chem. 1988, 263, 3411–3417. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Beuchat, M.H.; Lindsay, M.; Frias, S.; Palmiter, R.D.; Sakuraba, H.; Parton, R.G.; Gruenberg, J. Late Endosomal Membranes Rich in Lysobisphosphatidic Acid Regulate Cholesterol Transport. Nat. Cell Biol. 1999, 1, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Kirkegaard, T.; Gray, J.; Priestman, D.A.; Wallom, K.-L.; Atkins, J.; Olsen, O.D.; Klein, A.; Drndarski, S.; Petersen, N.H.T.; Ingemann, L.; et al. Heat Shock Protein-Based Therapy as a Potential Candidate for Treating the Sphingolipidoses. Sci. Transl. Med. 2016, 8, 355ra118. [Google Scholar] [CrossRef] [PubMed]
- Mengel, E.; Patterson, M.C.; Da Riol, R.M.; Del Toro, M.; Deodato, F.; Gautschi, M.; Grunewald, S.; Grønborg, S.; Harmatz, P.; Héron, B.; et al. Efficacy and Safety of Arimoclomol in Niemann-Pick Disease Type C: Results from a Double-Blind, Randomised, Placebo-Controlled, Multinational Phase 2/3 Trial of a Novel Treatment. J. Inherit. Metab. Dis. 2021, 44, 1463–1480. [Google Scholar] [CrossRef] [PubMed]
- Schedin-Weiss, S.; Winblad, B.; Tjernberg, L.O. The Role of Protein Glycosylation in Alzheimer Disease. FEBS J. 2014, 281, 46–62. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Yu, W.H.; Kumar, A.; Lee, S.; Mohan, P.S.; Peterhoff, C.M.; Wolfe, D.M.; Martinez-Vicente, M.; Massey, A.C.; Sovak, G.; et al. Lysosomal Proteolysis and Autophagy Require Presenilin 1 and Are Disrupted by Alzheimer-Related PS1 Mutations. Cell 2010, 141, 1146–1158. [Google Scholar] [CrossRef] [PubMed]
- Coen, K.; Flannagan, R.S.; Baron, S.; Carraro-Lacroix, L.R.; Wang, D.; Vermeire, W.; Michiels, C.; Munck, S.; Baert, V.; Sugita, S.; et al. Lysosomal Calcium Homeostasis Defects, Not Proton Pump Defects, Cause Endo-Lysosomal Dysfunction in PSEN-Deficient Cells. J. Cell Biol. 2012, 198, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Garbett, K.; Veeraraghavalu, K.; Wilburn, B.; Gilmore, R.; Mirnics, K.; Sisodia, S.S. A Role for Presenilins in Autophagy Revisited: Normal Acidification of Lysosomes in Cells Lacking PSEN1 and PSEN2. J. Neurosci. 2012, 32, 8633–8648. [Google Scholar] [CrossRef] [PubMed]
- Suga, K.; Tomiyama, T.; Mori, H.; Akagawa, K. Syntaxin 5 Interacts with Presenilin Holoproteins, but Not with Their N- or C-Terminal Fragments, and Affects Beta-Amyloid Peptide Production. Biochem. J. 2004, 381, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Suga, K.; Saito, A.; Tomiyama, T.; Mori, H.; Akagawa, K. Syntaxin 5 Interacts Specifically with Presenilin Holoproteins and Affects Processing of betaAPP in Neuronal Cells. J. Neurochem. 2005, 94, 425–439. [Google Scholar] [CrossRef] [PubMed]
- Linders, P.T.A.; Gerretsen, E.C.F.; Ashikov, A.; Vals, M.-A.; de Boer, R.; Revelo, N.H.; Arts, R.; Baerenfaenger, M.; Zijlstra, F.; Huijben, K.; et al. Congenital Disorder of Glycosylation Caused by Starting Site-Specific Variant in Syntaxin-5. Nat. Commun. 2021, 12, 6227. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, J.V.; Twomey, C.; Wujek, P. Presenilin-Dependent Regulated Intramembrane Proteolysis and Gamma-Secretase Activity. Cell Mol. Life Sci. 2009, 66, 1534–1555. [Google Scholar] [CrossRef] [PubMed]
- Kitano, M.; Kizuka, Y.; Sobajima, T.; Nakano, M.; Nakajima, K.; Misaki, R.; Itoyama, S.; Harada, Y.; Harada, A.; Miyoshi, E.; et al. Rab11-Mediated Post-Golgi Transport of the Sialyltransferase ST3GAL4 Suggests a New Mechanism for Regulating Glycosylation. J. Biol. Chem. 2021, 296, 100354. [Google Scholar] [CrossRef] [PubMed]
- Weissmiller, A.M.; Natera-Naranjo, O.; Reyna, S.M.; Pearn, M.L.; Zhao, X.; Nguyen, P.; Cheng, S.; Goldstein, L.S.B.; Tanzi, R.E.; Wagner, S.L.; et al. A γ-Secretase Inhibitor, but Not a γ-Secretase Modulator, Induced Defects in BDNF Axonal Trafficking and Signaling: Evidence for a Role for APP. PLoS ONE 2015, 10, e0118379. [Google Scholar] [CrossRef] [PubMed]
- Soto-Faguás, C.M.; Sanchez-Molina, P.; Saura, C.A. Loss of Presenilin Function Enhances Tau Phosphorylation and Aggregation in Mice. Acta Neuropathol. Commun. 2021, 9, 162. [Google Scholar] [CrossRef] [PubMed]
- Runz, H.; Rietdorf, J.; Tomic, I.; de Bernard, M.; Beyreuther, K.; Pepperkok, R.; Hartmann, T. Inhibition of Intracellular Cholesterol Transport Alters Presenilin Localization and Amyloid Precursor Protein Processing in Neuronal Cells. J. Neurosci. 2002, 22, 1679–1689. [Google Scholar] [CrossRef] [PubMed]
- Burns, M.; Gaynor, K.; Olm, V.; Mercken, M.; LaFrancois, J.; Wang, L.; Mathews, P.M.; Noble, W.; Matsuoka, Y.; Duff, K. Presenilin Redistribution Associated with Aberrant Cholesterol Transport Enhances Beta-Amyloid Production in Vivo. J. Neurosci. 2003, 23, 5645–5649. [Google Scholar] [CrossRef] [PubMed]
- Malnar, M.; Kosicek, M.; Mitterreiter, S.; Omerbasic, D.; Lichtenthaler, S.F.; Goate, A.; Hecimovic, S. Niemann-Pick Type C Cells Show Cholesterol Dependent Decrease of APP Expression at the Cell Surface and Its Increased Processing through the Beta-Secretase Pathway. Biochim. Biophys. Acta 2010, 1802, 682–691. [Google Scholar] [CrossRef]
- Lin, T.; van Husen, L.S.; Yu, Y.; Tjernberg, L.O.; Schedin-Weiss, S. Lack of N-Glycosylation Increases Amyloidogenic Processing of the Amyloid Precursor Protein. Glycobiology 2022, 32, 506–517. [Google Scholar] [CrossRef]
- Tienari, P.J.; De Strooper, B.; Ikonen, E.; Simons, M.; Weidemann, A.; Czech, C.; Hartmann, T.; Ida, N.; Multhaup, G.; Masters, C.L.; et al. The Beta-Amyloid Domain Is Essential for Axonal Sorting of Amyloid Precursor Protein. EMBO J. 1996, 15, 5218–5229. [Google Scholar] [CrossRef] [PubMed]
- Urano, Y.; Takahachi, M.; Higashiura, R.; Fujiwara, H.; Funamoto, S.; Imai, S.; Futai, E.; Okuda, M.; Sugimoto, H.; Noguchi, N. Curcumin Derivative GT863 Inhibits Amyloid-Beta Production via Inhibition of Protein N-Glycosylation. Cells 2020, 9, 349. [Google Scholar] [CrossRef]
- Habchi, J.; Chia, S.; Galvagnion, C.; Michaels, T.C.T.; Bellaiche, M.M.J.; Ruggeri, F.S.; Sanguanini, M.; Idini, I.; Kumita, J.R.; Sparr, E.; et al. Cholesterol Catalyses Aβ42 Aggregation through a Heterogeneous Nucleation Pathway in the Presence of Lipid Membranes. Nat. Chem. 2018, 10, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Kakio, A.; Nishimoto, S.I.; Yanagisawa, K.; Kozutsumi, Y.; Matsuzaki, K. Cholesterol-Dependent Formation of GM1 Ganglioside-Bound Amyloid Beta-Protein, an Endogenous Seed for Alzheimer Amyloid. J. Biol. Chem. 2001, 276, 24985–24990. [Google Scholar] [CrossRef]
- Hayashi, H.; Kimura, N.; Yamaguchi, H.; Hasegawa, K.; Yokoseki, T.; Shibata, M.; Yamamoto, N.; Michikawa, M.; Yoshikawa, Y.; Terao, K.; et al. A Seed for Alzheimer Amyloid in the Brain. J. Neurosci. 2004, 24, 4894–4902. [Google Scholar] [CrossRef] [PubMed]
- Yahi, N.; Aulas, A.; Fantini, J. How Cholesterol Constrains Glycolipid Conformation for Optimal Recognition of Alzheimer’s Beta Amyloid Peptide (Abeta1-40). PLoS ONE 2010, 5, e9079. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, T.; Chang, T.Y.; Haass, C.; Ihara, Y. Accumulation and Aggregation of Amyloid Beta-Protein in Late Endosomes of Niemann-Pick Type C Cells. J. Biol. Chem. 2001, 276, 4454–4460. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Suzuki, K.; Nanba, E.; Yamamoto, T.; Ohno, K.; Murayama, S. Niemann-Pick Type C Disease: Accelerated Neurofibrillary Tangle Formation and Amyloid Beta Deposition Associated with Apolipoprotein E Epsilon 4 Homozygosity. Ann. Neurol. 2002, 52, 351–355. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.-W.; Shie, F.-S.; Maezawa, I.; Vincent, I.; Bird, T. Intracellular Accumulation of Amyloidogenic Fragments of Amyloid-Beta Precursor Protein in Neurons with Niemann-Pick Type C Defects Is Associated with Endosomal Abnormalities. Am. J. Pathol. 2004, 164, 975–985. [Google Scholar] [CrossRef] [PubMed]
- Farfara, D.; Trudler, D.; Segev-Amzaleg, N.; Galron, R.; Stein, R.; Frenkel, D. γ-Secretase Component Presenilin Is Important for Microglia β-Amyloid Clearance. Ann. Neurol. 2011, 69, 170–180. [Google Scholar] [CrossRef]
- Glebov, K.; Wunderlich, P.; Karaca, I.; Walter, J. Functional Involvement of γ-Secretase in Signaling of the Triggering Receptor Expressed on Myeloid Cells-2 (TREM2). J. Neuroinflammation 2016, 13, 17. [Google Scholar] [CrossRef] [PubMed]
- Walter, J.; Kemmerling, N.; Wunderlich, P.; Glebov, K. γ-Secretase in Microglia—Implications for Neurodegeneration and Neuroinflammation. J. Neurochem. 2017, 143, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Hou, P.; Zielonka, M.; Serneels, L.; Martinez-Muriana, A.; Fattorelli, N.; Wolfs, L.; Poovathingal, S.; T’Syen, D.; Balusu, S.; Theys, T.; et al. The γ-Secretase Substrate Proteome and Its Role in Cell Signaling Regulation. Mol. Cell 2023, 83, 4106–4122.e10. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.; Sun, Y.; Gao, Y.; Nakamura, T.; Noorani, A.A.; Li, T.; Wong, P.C.; Kimura, N.; Matsubara, E.; Kasuga, K.; et al. Presenilin Is Essential for ApoE Secretion, a Novel Role of Presenilin Involved in Alzheimer’s Disease Pathogenesis. J. Neurosci. 2022, 42, 1574–1586. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Tandon, A.; Sanjo, N.; Gu, Y.-J.; Hasegawa, H.; Arawaka, S.; Lee, F.J.S.; Ruan, X.; Mastrangelo, P.; Erdebil, S.; et al. Presenilin 1 and Presenilin 2 Have Differential Effects on the Stability and Maturation of Nicastrin in Mammalian Brain. J. Biol. Chem. 2003, 278, 19974–19979. [Google Scholar] [CrossRef] [PubMed]
- Guttenplan, K.A.; Weigel, M.K.; Prakash, P.; Wijewardhane, P.R.; Hasel, P.; Rufen-Blanchette, U.; Münch, A.E.; Blum, J.A.; Fine, J.; Neal, M.C.; et al. Neurotoxic Reactive Astrocytes Induce Cell Death via Saturated Lipids. Nature 2021, 599, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Marschallinger, J.; Iram, T.; Zardeneta, M.; Lee, S.E.; Lehallier, B.; Haney, M.S.; Pluvinage, J.V.; Mathur, V.; Hahn, O.; Morgens, D.W.; et al. Lipid-Droplet-Accumulating Microglia Represent a Dysfunctional and Proinflammatory State in the Aging Brain. Nat. Neurosci. 2020, 23, 194–208. [Google Scholar] [CrossRef] [PubMed]
- Haney, M.S.; Pálovics, R.; Munson, C.N.; Long, C.; Johansson, P.K.; Yip, O.; Dong, W.; Rawat, E.; West, E.; Schlachetzki, J.C.M.; et al. APOE4/4 Is Linked to Damaging Lipid Droplets in Alzheimer’s Disease Microglia. Nature 2024, 628, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Kizuka, Y.; Kitazume, S.; Taniguchi, N. N-Glycan and Alzheimer’s Disease. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 2447–2454. [Google Scholar] [CrossRef] [PubMed]
- Haukedal, H.; Freude, K.K. Implications of Glycosylation in Alzheimer’s Disease. Front. Neurosci. 2020, 14, 625348. [Google Scholar] [CrossRef] [PubMed]
- Millard, E.E.; Srivastava, K.; Traub, L.M.; Schaffer, J.E.; Ory, D.S. Niemann-Pick Type C1 (NPC1) Overexpression Alters Cellular Cholesterol Homeostasis. J. Biol. Chem. 2000, 275, 38445–38451. [Google Scholar] [CrossRef] [PubMed]
- Fujitani, N.; Furukawa, J.; Araki, K.; Fujioka, T.; Takegawa, Y.; Piao, J.; Nishioka, T.; Tamura, T.; Nikaido, T.; Ito, M.; et al. Total Cellular Glycomics Allows Characterizing Cells and Streamlining the Discovery Process for Cellular Biomarkers. Proc. Natl. Acad. Sci. USA 2013, 110, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Kondo, K.; Harada, Y.; Nakano, M.; Suzuki, T.; Fukushige, T.; Hanzawa, K.; Yagi, H.; Takagi, K.; Mizuno, K.; Miyamoto, Y.; et al. Identification of Distinct N-Glycosylation Patterns on Extracellular Vesicles from Small-Cell and Non-Small-Cell Lung Cancer Cells. J. Biol. Chem. 2022, 298, 101950. [Google Scholar] [CrossRef] [PubMed]
- Uematsu, R.; Furukawa, J.; Nakagawa, H.; Shinohara, Y.; Deguchi, K.; Monde, K.; Nishimura, S.-I. High Throughput Quantitative Glycomics and Glycoform-Focused Proteomics of Murine Dermis and Epidermis. Mol. Cell Proteom. 2005, 4, 1977–1989. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, J.; Shinohara, Y.; Kuramoto, H.; Miura, Y.; Shimaoka, H.; Kurogochi, M.; Nakano, M.; Nishimura, S.-I. Comprehensive Approach to Structural and Functional Glycomics Based on Chemoselective Glycoblotting and Sequential Tag Conversion. Anal. Chem. 2008, 80, 1094–1101. [Google Scholar] [CrossRef] [PubMed]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fabiano, M.; Oikawa, N.; Kerksiek, A.; Furukawa, J.-i.; Yagi, H.; Kato, K.; Schweizer, U.; Annaert, W.; Kang, J.; Shen, J.; et al. Presenilin Deficiency Results in Cellular Cholesterol Accumulation by Impairment of Protein Glycosylation and NPC1 Function. Int. J. Mol. Sci. 2024, 25, 5417. https://doi.org/10.3390/ijms25105417
Fabiano M, Oikawa N, Kerksiek A, Furukawa J-i, Yagi H, Kato K, Schweizer U, Annaert W, Kang J, Shen J, et al. Presenilin Deficiency Results in Cellular Cholesterol Accumulation by Impairment of Protein Glycosylation and NPC1 Function. International Journal of Molecular Sciences. 2024; 25(10):5417. https://doi.org/10.3390/ijms25105417
Chicago/Turabian StyleFabiano, Marietta, Naoto Oikawa, Anja Kerksiek, Jun-ichi Furukawa, Hirokazu Yagi, Koichi Kato, Ulrich Schweizer, Wim Annaert, Jongkyun Kang, Jie Shen, and et al. 2024. "Presenilin Deficiency Results in Cellular Cholesterol Accumulation by Impairment of Protein Glycosylation and NPC1 Function" International Journal of Molecular Sciences 25, no. 10: 5417. https://doi.org/10.3390/ijms25105417





