Multi-Omics Reveals Disrupted Immunometabolic Homeostasis and Oxidative Stress in Adipose Tissue of Dairy Cows with Subclinical Ketosis: A Sphingolipid-Centric Perspective
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design, Retrospective Analysis, and Animal Management
2.2. Sampling
2.3. Serum Metabolic Status Parameters Analysis
2.4. Biochemical Parameters Analysis of Subcutaneous Adipose Tissue
2.5. Whole Blood and Adipose Tissue RNA-Seq Data Analysis
2.6. Quantitative RT-PCR
2.7. Serum and Adipose Tissue Untargeted Lipidome Analysis
2.8. Targeted Lipidome for Sphingolipids in Adipose Tissue
2.9. Statistical Analysis
3. Results
3.1. Serum Metabolic Status Parameters
3.2. Biochemical Parameters of Subcutaneous Adipose Tissue
3.3. Whole Blood and Adipose Tissue Transcriptome
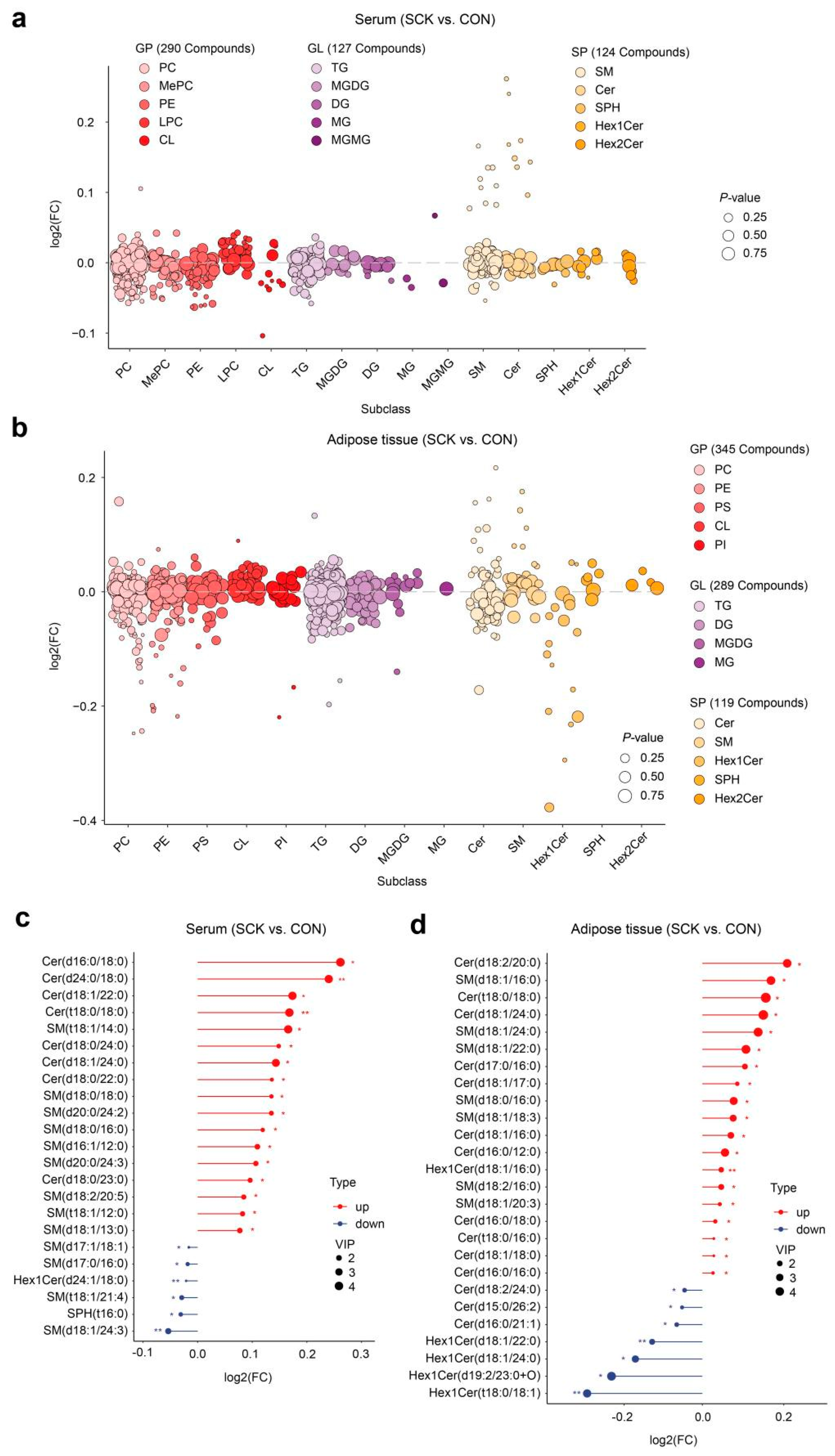
3.4. Serum and Adipose Tissue Lipidome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grummer, R.R.; Mashek, D.G.; Hayirli, A. Dry matter intake and energy balance in the transition period. Vet. Clin. N. Am. Food Anim. Pract. 2004, 20, 447–470. [Google Scholar] [CrossRef] [PubMed]
- Drackley, J.K. Biology of dairy cows during the transition period: The final frontier? J. Dairy. Sci. 1999, 82, 2259–2273. [Google Scholar] [CrossRef]
- Herdt, T.H. Ruminant adaptation to negative energy balance—Influences on the etiology of ketosis and fatty liver. Vet. Clin. N. Am. Food Anim. Pract. 2000, 16, 215–230. [Google Scholar] [CrossRef]
- Opsomer, G. Interaction between metabolic challenges and productivity in high yielding dairy cows. Jpn. J. Vet. Res. 2015, 63 (Suppl. 1), S1–S14. [Google Scholar] [PubMed]
- Andersson, L. Subclinical Ketosis in Dairy Cows. Vet. Clin. N. Am. Food Anim. Pract. 1988, 4, 233–251. [Google Scholar] [CrossRef]
- Esposito, G.; Irons, P.C.; Webb, E.C.; Chapwanya, A. Interactions between negative energy balance, metabolic diseases, uterine health and immune response in transition dairy cows. Anim. Reprod. Sci. 2014, 144, 60–71. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, L.; Xu, C.; Xia, C.; Sun, L.; Shu, S. Plasma metabolomic profiling of dairy cows affected with ketosis using gas chromatography/mass spectrometry. BMC Vet. Res. 2013, 9, 186. [Google Scholar] [CrossRef]
- Li, Y.; Xu, C.; Xia, C.; Zhang, H.; Sun, L.; Gao, Y. Plasma metabolic profiling of dairy cows affected with clinical ketosis using LC/MS technology. Vet. Quart. 2014, 34, 152–158. [Google Scholar] [CrossRef]
- Shahzad, K.; Lopreiato, V.; Liang, Y.; Trevisi, E.; Osorio, J.S.; Xu, C.; Loor, J.J. Hepatic metabolomics and transcriptomics to study susceptibility to ketosis in response to prepartal nutritional management. J. Anim. Sci. Biotechnol. 2019, 10, 96. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.C.; Zhao, H.Y.; Li, L.X.; Tan, J.; Wang, Y.; Liu, M.; Jiang, L.S. Multi-omics analysis reveals that the metabolite profile of raw milk is associated with dairy cows’ health status. Food Chem. 2023, 428, 136813. [Google Scholar] [CrossRef] [PubMed]
- Kershaw, E.E.; Flier, J.S. Adipose tissue as an endocrine organ. J. Clin. Endocrinol. Metab. 2004, 89, 2548–2556. [Google Scholar] [CrossRef] [PubMed]
- Pravettoni, D.; Doll, K.; Hummel, M.; Cavallone, E.; Re, M.; Belloli, A.G. Insulin resistance and abomasal motility disorders in cows detected by use of abomasoduodenal electromyography after surgical correction of left displaced abomasum. Am. J. Vet. Res. 2004, 65, 1319–1324. [Google Scholar] [CrossRef] [PubMed]
- De Koster, J.D.; Opsomer, G. Insulin resistance in dairy cows. Vet. Clin. N. Am. Food Anim. Pract. 2013, 29, 299–322. [Google Scholar] [CrossRef] [PubMed]
- Rocco, S.M.; McNamara, J.P. Regulation of bovine adipose tissue metabolism during lactation. 7. Metabolism and gene expression as a function of genetic merit and dietary energy intake. J. Dairy. Sci. 2013, 96, 3108–3119. [Google Scholar] [CrossRef] [PubMed]
- Mellouk, N.; Rame, C.; Naquin, D.; Jaszczyszyn, Y.; Touzé, J.L.; Briant, E.; Guillaume, D.; Ntallaris, T.; Humblot, P.; Dupont, J. Impact of the severity of negative energy balance on gene expression in the subcutaneous adipose tissue of periparturient primiparous Holstein dairy cows: Identification of potential novel metabolic signals for the reproductive system. PLoS ONE 2019, 14, e0222954. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Li, X.; Ma, L.; Loor, J.J.; Coleman, D.N.; Jia, H.; Liu, G.; Xu, C.; Wang, Y.; Li, X.W. Adipose tissue proteomic analysis in ketotic or healthy Holstein cows in early lactation. J. Anim. Sci. 2019, 97, 2837–2849. [Google Scholar] [CrossRef]
- Ning, M.; Zhao, Y.; Dai, D.; Yao, C.; Liu, H.; Fang, L.; Wang, B.; Zhang, Y.; Cao, J. Gene co-expression network and differential expression analyses of subcutaneous white adipose tissue reveal novel insights into the pathological mechanisms underlying ketosis in dairy cows. J. Dairy. Sci. 2023, 106, 5018–5028. [Google Scholar] [CrossRef] [PubMed]
- Loizides-Mangold, U. On the future of mass-spectrometry-based lipidomics. FEBS J. 2013, 280, 2817–2829. [Google Scholar] [CrossRef] [PubMed]
- Gerspach, C.; Imhasly, S.; Gubler, M.; Naegeli, H.; Ruetten, M.; Laczko, E. Altered plasma lipidome profile of dairy cows with fatty liver disease. Res. Vet. Sci. 2017, 110, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Toma, R.; Duval, N.; Pelle, B.; Parks, M.M.; Gopu, V.; Torres, P.J.; Camacho, F.R.; Shen, N.; Krishnan, S.; Hatch, A.; et al. A clinically validated human capillary blood transcriptome test for global systems biology studies. BioTechniques 2020, 69, 289–301. [Google Scholar] [CrossRef]
- Chaussabel, D.; Pascual, V.; Banchereau, J. Assessing the human immune system through blood transcriptomics. BMC Biol. 2010, 8, 84. [Google Scholar] [CrossRef] [PubMed]
- NRC. Nutrient Requirements of Dairy Cattle, 7th ed.; National Research Council: Washington, DC, USA, 2001.
- Association of Official Analytical Chemists (AOAC). Official Methods of Analysis; Association of Official Analytical Chemists: Gaithersburg, MD, USA, 2012. [Google Scholar]
- Van Soest, P.J.; Robertson, J.B.; Lewis, B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy. Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef] [PubMed]
- Holtenius, P.; Holtenius, K. A model to estimate insulin sensitivity in dairy cows. Acta Vet. Scand. 2007, 49, 29. [Google Scholar] [CrossRef] [PubMed]
- Vailati-Riboni, M.; Farina, G.; Batistel, F.; Heiser, A.; Mitchell, M.D.; Crookenden, M.A.; Walker, C.G.; Kay, J.K.; Meier, S.; Roche, J.R.; et al. Far-off and close-up dry matter intake modulate indicators of immunometabolic adaptations to lactation in subcutaneous adipose tissue of pasture-based transition dairy cows. J. Dairy. Sci. 2017, 100, 2334–2350. [Google Scholar] [CrossRef] [PubMed]
- Vailati-Riboni, M.; Kanwal, M.; Bulgari, O.; Meier, S.; Priest, N.V.; Burke, C.R.; Kay, J.K.; McDougall, S.; Mitchell, M.D.; Walker, C.G.; et al. Body condition score and plane of nutrition prepartum affect adipose tissue transcriptome regulators of metabolism and inflammation in grazing dairy cows during the transition period. J. Dairy. Sci. 2016, 99, 758–770. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Alharthi, A.S.; Bucktrout, R.; Elolimy, A.A.; Lopreiato, V.; Martinez-Cortés, I.; Xu, C.; Fernandez, C.; Trevisi, E.; Loor, J.J. Body condition alters glutathione and nuclear factor erythroid 2-like 2 (NFE2L2)- related antioxidant network abundance in subcutaneous adipose tissue of periparturient Holstein cows. J. Dairy. Sci. 2020, 103, 6439–6453. [Google Scholar] [CrossRef] [PubMed]
- Draijer, L.G.; Froon-Torenstra, D.; van Weeghel, M.; Vaz, F.M.; Bohte, A.E.; Holleboom, A.G.; Benninga, M.A.; Koot, B.G.P. Lipidomics in nonalcoholic fatty liver disease: Exploring serum lipids as biomarkers for pediatric nonalcoholic fatty liver disease. J. Pediatr. Gastroenterol. Nutr. 2020, 71, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Chaves-Filho, A.B.; Peixoto, A.S.; Castro, Ã.; Oliveira, T.E.; Perandini, L.A.; Moreira, R.J.; Silva, R.P.; Silva, B.P.; Moretti, E.H.; Steiner, A.A.; et al. Futile cycle of β-oxidation and de novo lipogenesis are associated with essential fatty acids depletion in lipoatrophy. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2023, 1868, 159264. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Báez, J.; Risco, C.A.; Chebel, R.C.; Gomes, G.C.; Greco, L.F.; Tao, S.; Thompson, I.M.; Amaral, B.C.; Zenobi, M.G.; Martinez, N.; et al. Association of dry matter intake and energy balance prepartum and postpartum with health disorders postpartum: Part II. Ketosis and clinical mastitis. J. Dairy. Sci. 2019, 102, 9151–9164. [Google Scholar] [CrossRef] [PubMed]
- Kehrli, J.M.; Neil, J.; Burvenich, C.; Goff, J.; Lippolis, J.; Reinhardt, T.; Nonnecke, B. Ruminant Physiology: Digestion, Metabolism and Impact of Nutrition on Gene Expression, Immunology and Stress. In Energy and Protein Effects on the Immune System; Wageningen Academic Publishers: Wageningen, The Netherlands, 2006; pp. 455–471. [Google Scholar]
- Zu, L.; He, J.; Jiang, H.; Xu, C.; Pu, S.; Xu, G. Bacterial endotoxin stimulates adipose lipolysis via Toll-like receptor 4 and extracellular signal-regulated kinase pathway. J. Biol. Chem. 2009, 284, 5915–5926. [Google Scholar] [CrossRef]
- Arner, P.; Langin, D. Lipolysis in lipid turnover, cancer cachexia, and obesity-induced insulin resistance. Trends Endocrinol. Metab. 2014, 25, 255–262. [Google Scholar] [CrossRef]
- Chirivi, M.; Cortes, D.; Rendon, C.J.; Contreras, G.A. Lipolysis inhibition as a treatment of clinical ketosis in dairy cows: Effects on adipose tissue metabolic and immune responses. J. Dairy. Sci. 2024, in press. [Google Scholar] [CrossRef]
- Pires, J.A.A.; Souza, A.H.; Grummer, R.R. Induction of hyperlipidemia by intravenous infusion of tallow emulsion causes insulin resistance in holstein cows. J. Dairy. Sci. 2007, 90, 2735–2744. [Google Scholar] [CrossRef]
- Li, M.; Chi, X.; Wang, Y.; Setrerrahmane, S.; Xie, W.; Xu, H. Trends in insulin resistance: Insights into mechanisms and therapeutic strategy. Signal Transduct. Target. Ther. 2022, 7, 216. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.N.; Myers, W.A.; Eduardo Rico, J.; Feng, W.L.; Chang, C.; Richards, A.T.; Moniruzzaman, M.; Haughey, N.J.; McFadden, J.W. Effects of serine palmitoyltransferase inhibition by myriocin in ad libitum-fed and nutrient-restricted ewes. J. Anim. Sci. 2021, 99, skab221. [Google Scholar]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Siow, D.L.; Wattenberg, B.W. Mammalian ORMDL proteins mediate the feedback response in ceramide biosynthesis. J. Biol. Chem. 2012, 287, 40198–40204. [Google Scholar] [CrossRef]
- Hannun, Y.A.; Obeid, L.M. Principles of bioactive lipid signalling: Lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 2008, 9, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Rico, J.E.; Bandaru, V.V.; Dorskind, J.M.; Haughey, N.J.; McFadden, J.W. Plasma ceramides are elevated in overweight Holstein dairy cows experiencing greater lipolysis and insulin resistance during the transition from late pregnancy to early lactation. J. Dairy. Sci. 2015, 98, 7757–7770. [Google Scholar] [CrossRef]
- Davis, A.N.; Clegg, J.L.; Perry, C.A.; McFadden, J.W. Nutrient restriction increases circulating and hepatic ceramide in dairy cows displaying impaired insulin tolerance. Lipids 2017, 52, 771–780. [Google Scholar] [CrossRef]
- Kenez, A.; Bassler, S.C.; Jorge-Smeding, E.; Huber, K. Ceramide metabolism associated with chronic dietary nutrient surplus and diminished insulin sensitivity in the liver, muscle, and adipose tissue of cattle. Front. Physiol. 2022, 13, 958837. [Google Scholar] [CrossRef]
- Xu, C.; Shu, S.; Xia, C.; Wang, B.; Zhang, H.; Jun, B. Investigation on the relationship of insulin resistance and ketosis in dairy cows. J. Vet. Sci. Technol. 2014, 5, 62. [Google Scholar]
- Youssef, M.A.; El-Ashker, M.R.; Younis, M.S. The effect of subclinical ketosis on indices of insulin sensitivity and selected metabolic variables in transition dairy cattle. Comp. Clin. Pathol. 2017, 26, 329–334. [Google Scholar] [CrossRef]
- McFadden, J.W.; Rico, J.E. Invited review: Sphingolipid biology in the dairy cow: The emerging role of ceramide. J. Dairy. Sci. 2019, 102, 7619–7639. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhao, H.; Li, L.; Yu, S.; Liu, M.; Jiang, L. Ceramide on the road to insulin resistance and immunometabolic disorders in transition dairy cows: Driver or passenger? Front. Immunol. 2024, 14, 1321597. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.N.; Myers, W.A.; Chang, C.; Tate, B.N.; Rico, J.E.; Moniruzzaman, M.; Haughey, N.J.; McFadden, J.W. Somatotropin increases plasma ceramide in relation to enhanced milk yield in cows. Domest. Anim. Endocrinol. 2021, 74, 106480. [Google Scholar] [CrossRef] [PubMed]
- Rico, J.E.; Myers, W.A.; Laub, D.J.; Davis, A.N.; Zeng, Q.; McFadden, J.W. Hot topic: Ceramide inhibits insulin sensitivity in primary bovine adipocytes. J. Dairy. Sci. 2018, 101, 3428–3432. [Google Scholar] [CrossRef] [PubMed]
- Chavez, J.A.; Summers, S.A. A Ceramide-Centric View of Insulin Resistance. Cell Metab. 2012, 15, 585–594. [Google Scholar] [CrossRef]
- Hotta, K.; Funahashi, T.; Bodkin, N.L.; Ortmeyer, H.K.; Arita, Y.; Hansen, B.C.; Matsuzawa, Y. Circulating concentrations of the adipocyte protein adiponectin are decreased in parallel with reduced insulin sensitivity during the progression to Type 2 diabetes in rhesus monkeys. Diabetes 2001, 50, 1126–1133. [Google Scholar] [CrossRef]
- Fasshauer, M.; Klein, J.; Neumann, S.; Eszlinger, M.; Paschke, R. Adiponectin gene expression is inhibited by β-adrenergic stimulation via protein kinase A in 3T3-L1 adipocytes. FEBS Lett. 2001, 507, 142–146. [Google Scholar] [CrossRef]
- Akgul, G.; Mecitoglu, Z.; Kucuksen, D.U.; Senturk, S. Comparison of adiponectin levels and some metabolic parameters in dairy cows with subclinical and clinical ketosis. Med. Weter.-Vet. Med.-Sci. Pract. 2018, 74, 182–186. [Google Scholar] [CrossRef]
- Mann, S.; Urh, C.; Sauerwein, H.; Wakshlag, J.J.; Yepes, F.A.L.; Overton, T.R.; Nydam, D.V. Short communication: The association of adiponectin and leptin concentrations with prepartum dietary energy supply, parity, body condition, and postpartum hyperketonemia in transition dairy cows. J. Dairy. Sci. 2018, 101, 806–811. [Google Scholar] [CrossRef]
- Kabara, E.; Sordillo, L.M.; Holcombe, S.; Contreras, G.A. Adiponectin links adipose tissue function and monocyte inflammatory responses during bovine metabolic stress. Comp. Immunol. Microbiol. Infect. Dis. 2014, 37, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, T.; Kamon, J.; Waki, H.; Terauchi, Y.; Kubota, N.; Hara, K.; Mori, Y.; Ide, T.; Murakami, K.; Tsuboyama-Kasaoka, N.; et al. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat. Med. 2001, 7, 941–946. [Google Scholar] [CrossRef] [PubMed]
- VasiliauskaitÃ-Brooks, I.; Sounier, R.; Rochaix, P.; Bellot, G.; Fortier, M.; Hoh, F.; Colibus, L.D.; Bechara, C.; Saied, E.M.; Arenz, C.; et al. Structural insights into adiponectin receptors suggest ceramidase activity. Nature 2017, 544, 120–123. [Google Scholar] [CrossRef] [PubMed]
- Al-Rashed, F.; Ahmad, Z.; Thomas, R.; Melhem, M.; Snider, A.J.; Obeid, L.M.; Al-Mulla, F.; Hannun, Y.A.; Ahmad, R. Neutral sphingomyelinase 2 regulates inflammatory responses in monocytes/macrophages induced by TNF-α. Sci. Rep. 2020, 10, 16802. [Google Scholar] [CrossRef] [PubMed]
- Meyer, S.G.E.; de Groot, H. Cycloserine and threo-dihydrosphingosine inhibit TNF-α-induced cytotoxicity: Evidence for the importance of de novo ceramide synthesis in TNF-α signaling. Biochim. Biophys. Acta 2003, 1643, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Yepes, M. Tissue-type plasminogen activator is a neuroprotectant in the central nervous system. Front. Cell. Neurosci. 2015, 9, 304. [Google Scholar] [CrossRef] [PubMed]
- Chimin, P.; Andrade, M.L.; Belchior, T.; Paschoal, V.A.; Magdalon, J.; Yamashita, A.S.; Castro, É.; Castoldi, A.; Chaves-Filho, A.B.; Yoshinaga, M.Y.; et al. Adipocyte mTORC1 deficiency promotes adipose tissue inflammation and NLRP3 inflammasome activation via oxidative stress and de novo ceramide synthesis. J. Lipid Res. 2017, 58, 1797–1807. [Google Scholar] [CrossRef] [PubMed]
- Scheiblich, H.; Schlütter, A.; Golenbock, D.T.; Latz, E.; Martinez-Martinez, P.; Heneka, M.T. Activation of the NLRP3 inflammasome in microglia: The role of ceramide. J. Neurochem. 2017, 143, 534–550. [Google Scholar] [CrossRef]
- Vandanmagsar, B.; Youm, Y.H.; Ravussin, A.; Galgani, J.E.; Stadler, K.; Mynatt, R.L.; Ravussin, E.; Stephens, J.M.; Dixit, V.D. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat. Med. 2011, 17, 179–188. [Google Scholar] [CrossRef]
- Kosteli, A.; Sugaru, E.; Haemmerle, G.; Martin, J.F.; Lei, J.; Zechner, R.; Ferrante, A.W., Jr. Weight loss and lipolysis promote a dynamic immune response in murine adipose tissue. J. Clin. Investig. 2010, 120, 3466–3479. [Google Scholar] [CrossRef] [PubMed]
- Contreras, G.A.; Thelen, K.; Schmidt, S.E.; Strieder-Barboza, C.; Preseault, C.L.; Raphael, W.; Kiupel, M.; Caron, J.; Lock, A.L. Adipose tissue remodeling in late-lactation dairy cows during feed-restriction-induced negative energy balance. J. Dairy. Sci. 2016, 99, 10009–10021. [Google Scholar] [CrossRef] [PubMed]
- Zachut, M.; Contreras, G.A. Symposium review: Mechanistic insights into adipose tissue inflammation and oxidative stress in periparturient dairy cows. J. Dairy. Sci. 2022, 105, 3670–3686. [Google Scholar] [CrossRef] [PubMed]
- Abuajamieh, M.; Kvidera, S.K.; Fernandez, M.V.; Nayeri, A.; Upah, N.C.; Nolan, E.A.; Lei, S.M.; DeFrain, J.M.; Green, H.B.; Schoenberg, K.M.; et al. Inflammatory biomarkers are associated with ketosis in periparturient Holstein cows. Res. Vet. Sci. 2016, 109, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Jiang, S.; Li, C.; Loor, J.J.; Jiang, Q.; Yang, Y.; Feng, X.; Liu, S.; He, J.; Wang, K.; et al. Free fatty acids promote degranulation of azurophil granules in neutrophils by inducing production of NADPH oxidase-derived reactive oxygen species in cows with subclinical ketosis. J. Dairy. Sci. 2022, 105, 2473–2486. [Google Scholar] [CrossRef] [PubMed]
- Chirivi, M.; Cortes-Beltran, D.; Munsterman, A.; O’Connor, A.; Contreras, G.A. Lipolysis inhibition as a treatment of clinical ketosis in dairy cows: A randomized clinical trial. J. Dairy. Sci. 2023, 106, 9514–9531. [Google Scholar] [CrossRef] [PubMed]
- Brodzki, P.; Marczuk, J.; Lisiecka, U.; Szczubia‚, M.; Brodzki, A.; GorzkoÅ, H.; Kulpa, K. Comparative evaluation of cytokine and acute-phase protein concentrations in sera of dairy cows with subclinical and clinical ketosis as a different view of the causes of the disease. Vet. World 2021, 14, 1572–1578. [Google Scholar] [CrossRef]
- Swartz, T.H.; Moallem, U.; Kamer, H.; Kra, G.; Levin, Y.; Mamedova, L.K.; Bradford, B.J.; Zachut, M. Characterization of the liver proteome in dairy cows experiencing negative energy balance at early lactation. J. Proteom. 2021, 246, 104308. [Google Scholar] [CrossRef]
- Brodzki, P.; Kostro, K.; Brodzki, A.; Wawron, W.; Marczuk, J.; Kurek, Å. Inflammatory cytokines and acute-phase proteins concentrations in the peripheral blood and uterus of cows that developed endometritis during early postpartum. Theriogenology 2015, 84, 11–18. [Google Scholar] [CrossRef]
- Castillo, C.; Hernández, J.; Valverde, I.; Pereira, V.; Sotillo, J.; Alonso, M.L.; Benedito, J.L. Plasma malonaldehyde (MDA) and total antioxidant status (TAS) during lactation in dairy cows. Res. Vet. Sci. 2006, 80, 133–139. [Google Scholar] [CrossRef]
- Castillo, C.; Hernandez, J.; Bravo, A.; Lopez-Alonso, M.; Pereira, V.; Benedito, J.L. Oxidative status during late pregnancy and early lactation in dairy cows. Vet. J. 2005, 169, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Li, N.; Gu, J.; Fu, S.; Peng, Z.; Zhao, C.; Zhang, Y.; Li, X.; Wang, Z.; Li, X.; et al. β-Hydroxybutyrate induces bovine hepatocyte apoptosis via an ROS-p38 signaling pathway. J. Dairy. Sci. 2016, 99, 9184–9198. [Google Scholar] [CrossRef] [PubMed]
- Senoh, T.; Oikawa, S.; Nakada, K.; Tagami, T.; Iwasaki, T. Increased serum malondialdehyde concentration in cows with subclinical ketosis. J. Vet. Med. Sci. 2019, 81, 817–820. [Google Scholar] [CrossRef] [PubMed]
- Garcà a-Ruiz, C.; Colell, A.; MarÃ, M.; Morales, A.; Fernández-Checa, J.C. Direct Effect of ceramide on the mitochondrial electron transport chain leads to generation of reactive oxygen species: Role of mitochondrial glutathione. J. Biol. Chem. 1997, 272, 11369–11377. [Google Scholar] [CrossRef] [PubMed]
- Rippo, M.R.; Malisan, F.; Rayagnan, L.; Tomassini, B.; Condo, I.; Costantini, P.; Susin, S.A.; Rufini, A.; Todaro, M.; Kroemer, G.; et al. GD3 ganglioside directly targets mitochondria in a bcl-2-controlled fashion. FASEB J. 2000, 14, 2047–2054. [Google Scholar] [CrossRef] [PubMed]
- Gudz, T.I.; Tserng, K.Y.; Hoppel, C.L. Direct inhibition of mitochondrial respiratory chain complex iii by cell-permeable ceramide. J. Biol. Chem. 1997, 272, 24154–24158. [Google Scholar] [CrossRef] [PubMed]
- Mason, S.D.; Rundqvist, H.; Papandreou, I.; Duh, R.; McNulty, W.J.; Howlett, R.A.; Olfert, I.M.; Sundberg, C.J.; Denko, N.C.; Poellinger, L.; et al. HIF-1α in endurance training: Suppression of oxidative metabolism. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007, 293, R2059–R2069. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Gao, Z.; Yin, J.; He, Q. Hypoxia is a potential risk factor for chronic inflammation and adiponectin reduction in adipose tissue of ob/ob and dietary obese mice. Am. J. Physiol. Endocrinol. Metab. 2007, 293, E1118–E1128. [Google Scholar] [CrossRef]
- Yin, J.; Gao, Z.; He, Q.; Zhou, D.; Guo, Z.; Ye, J. Role of hypoxia in obesity-induced disorders of glucose and lipid metabolism in adipose tissue. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E333–E342. [Google Scholar] [CrossRef]
- Wang, P.; Zeng, G.; Yan, Y.; Zhang, S.-y.; Dong, Y.; Zhang, Y.; Zhang, X.; Liu, H.; Zhang, Z.; Jiang, C.; et al. Disruption of adipocyte HIF-1α improves atherosclerosis through the inhibition of ceramide generation. Acta Pharm. Sin. B 2022, 12, 1899–1912. [Google Scholar] [CrossRef]
- Shen, T.; Li, X.; Loor, J.J.; Zhu, Y.; Du, X.; Wang, X.; Xing, D.; Shi, Z.; Fang, Z.; Li, X.; et al. Hepatic nuclear factor kappa B signaling pathway and NLR family pyrin domain containing 3 inflammasome is over-activated in ketotic dairy cows. J. Dairy. Sci. 2019, 102, 10554–10563. [Google Scholar] [CrossRef] [PubMed]
- Mohsin, M.A.; Yu, H.; He, R.; Wang, P.; Gan, L.; Du, Y.; Huang, Y.; Abro, M.B.; Sohaib, S.; Pierzchala, M.; et al. Differentiation of subclinical ketosis and liver function test indices in adipose tissues associated with hyperketonemia in postpartum dairy cattle. Front. Vet. Sci. 2021, 8, 796494. [Google Scholar] [CrossRef] [PubMed]
- Kenéz, Á.; Ruda, L.; Dänicke, S.; Huber, K. Insulin signaling and insulin response in subcutaneous and retroperitoneal adipose tissue in Holstein cows during the periparturient period. J. Dairy. Sci. 2019, 102, 11718–11729. [Google Scholar] [CrossRef] [PubMed]
- Ji, P.; Drackley, J.K.; Khan, M.J.; Loor, J.J. Inflammation- and lipid metabolism-related gene network expression in visceral and subcutaneous adipose depots of Holstein cows. J. Dairy. Sci. 2014, 97, 3441–3448. [Google Scholar] [CrossRef] [PubMed]
- Locher, L.F.; Meyer, N.; Weber, E.-M.; Rehage, J.; Meyer, U.; Dänicke, S.; Huber, K. Hormone-sensitive lipase protein expression and extent of phosphorylation in subcutaneous and retroperitoneal adipose tissues in the periparturient dairy cow. J. Dairy. Sci. 2011, 94, 4514–4523. [Google Scholar] [CrossRef] [PubMed]
- Leung, Y.H.; Christiane, B.S.; Koch, C.; Scheu, T.; Meyer, U.; Dänicke, S.; Huber, K.; Kenéz, Á. Sphingolipid profiling reveals different extent of ceramide accumulation in bovine retroperitoneal and subcutaneous adipose tissues. Metabolites 2020, 10, 473. [Google Scholar] [CrossRef]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, H.; Li, L.; Tan, J.; Wang, Y.; Zhang, A.; Zhao, Y.; Jiang, L. Multi-Omics Reveals Disrupted Immunometabolic Homeostasis and Oxidative Stress in Adipose Tissue of Dairy Cows with Subclinical Ketosis: A Sphingolipid-Centric Perspective. Antioxidants 2024, 13, 614. https://doi.org/10.3390/antiox13050614
Zhao H, Li L, Tan J, Wang Y, Zhang A, Zhao Y, Jiang L. Multi-Omics Reveals Disrupted Immunometabolic Homeostasis and Oxidative Stress in Adipose Tissue of Dairy Cows with Subclinical Ketosis: A Sphingolipid-Centric Perspective. Antioxidants. 2024; 13(5):614. https://doi.org/10.3390/antiox13050614
Chicago/Turabian StyleZhao, Huiying, Liuxue Li, Jian Tan, Ying Wang, Ao Zhang, Yuchao Zhao, and Linshu Jiang. 2024. "Multi-Omics Reveals Disrupted Immunometabolic Homeostasis and Oxidative Stress in Adipose Tissue of Dairy Cows with Subclinical Ketosis: A Sphingolipid-Centric Perspective" Antioxidants 13, no. 5: 614. https://doi.org/10.3390/antiox13050614






