Specific Cytotoxic Effects of Parasporal Crystal Proteins Isolated from Native Saudi Arabian Bacillus thuringiensis Strains against Cervical Cancer Cells
Abstract
:1. Introduction
2. Results
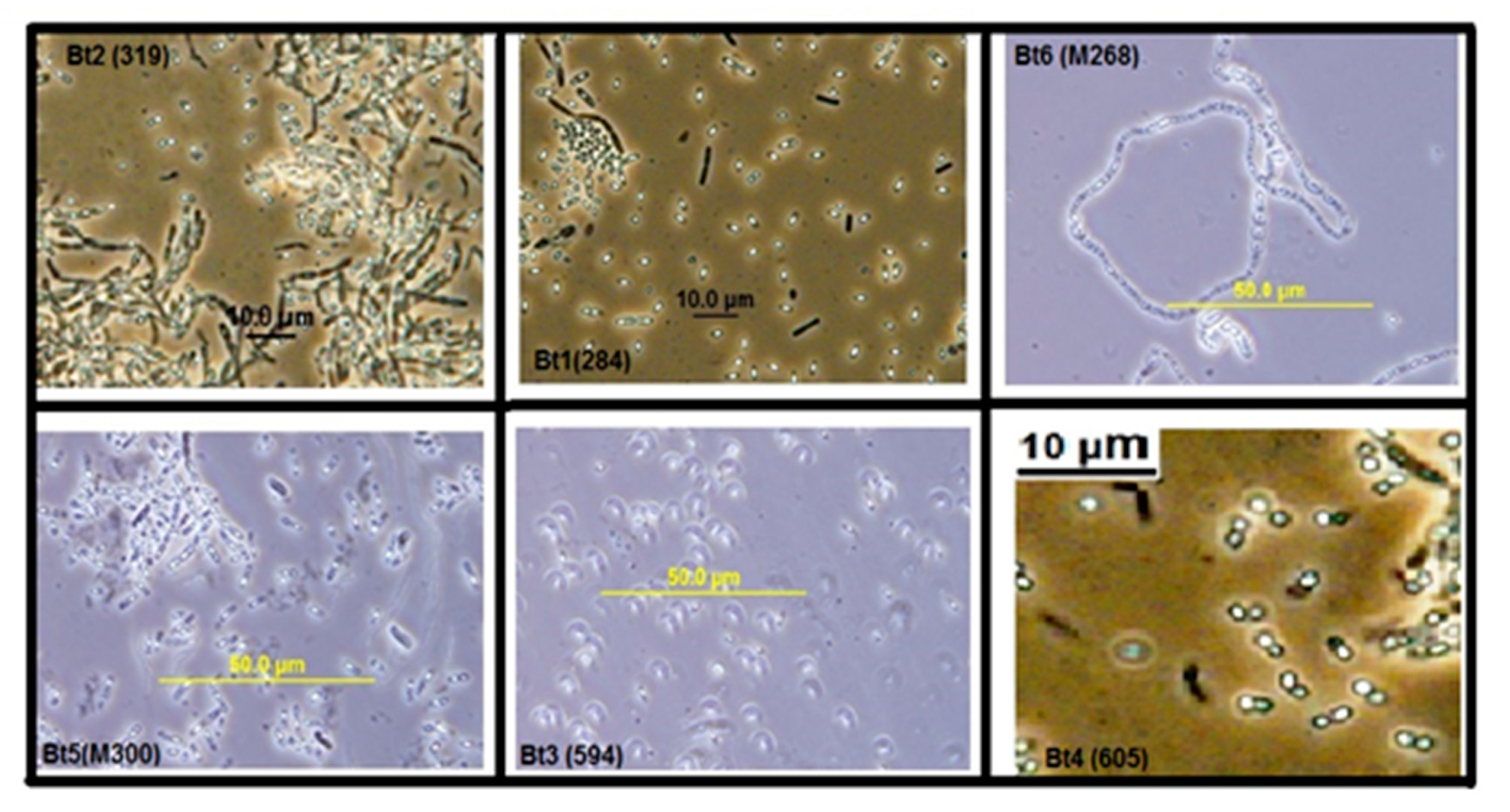
2.1. Phase Contrast and Electron Microscopy
2.2. Biochemical and 16S Molecular Typing of Isolated Bt Strains
2.3. PCR Analysis of Bt Isolates
2.4. SDS-PAGE Analysis of Nascent and Trypsin-Activated PS Proteins
2.5. Specific Anti-Proliferative Effects of PS Proteins
2.6. Transcriptional Activation of Apoptosis-Related Genes
3. Discussion
4. Materials and Methods
4.1. Ethical Approval
4.2. Chemical and Supplies
4.3. Isolation, Culturing and Identification of B. Thuringiensis Isolates
4.4. Biochemical and Molecular Typing
4.5. Scanning Electron Microscopy
4.6. Cell Lines and Culture Conditions
4.7. Hemolytic Assay
4.8. Crystal Sporal Mixture Isolation
4.9. Solubilization and Activation of the Crude Parasporal Crystal Proteins
4.10. PCR Screening of Bt Isolates for Parasporin (PS) Genes
4.11. Protein Chemistry
4.12. Measurement of Cellular Viability by MTT Assay
4.13. Gene Expression Profiling of Apoptotic Genes by Quantitative Reverse Transcriptase Polymerase Chain Reaction (RT-qPCR)
4.14. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- The International Agency for Research on Cancer (IARC), Global Cancer Observatory, World Health Organization (WHO). Report on the Latest Estimates on the Global Burden of Cancer. Date of Release: 12 September 2018 in Geneva, Switzerland. Available online: https://www.iarc.fr/wp-content/uploads/2018/09/pr263_E.pdf (accessed on 13 December 2018).
- Bazarbashi, S.; Al Eid, H.; Minguet, J. Cancer Incidence in Saudi Arabia: 2012 Data from the Saudi Cancer Registry. Asian Pac. J. Cancer Prev. 2017, 18, 2437–2444. [Google Scholar] [PubMed]
- Ibrahim, E.M.; Zeeneldin, A.A.; El-Khodary, T.R.; Al-Gahmi, A.M.; Bin Sadiq, B.M. Past, Present and Future of Colorectal Cancer in the Kingdom of Saudi Arabia. Saudi J. Gastroenterol. 2008, 14, 178–182. [Google Scholar] [CrossRef] [PubMed]
- Are, C.; Rajaram, S.; Are, M.; Raj, H.; Anderson, B.O.; Chaluvarya Swamy, R.; Vijayakumar, M.; Song, T.; Pandey, M.; Edney, J.A.; et al. A review of global cancer burden: Trends, challenges, strategies, and a role for surgeons. J. Surg. Oncol. 2013, 107, 221–226. [Google Scholar] [CrossRef]
- Cancer Council Australia. Understanding Chemotherapy: A Guide for People with Cancer, Their Families and Friends; Cancer Council Australia: Woolloomooloo, Australia, 2016; ISBN 978 1 925136 18 0. [Google Scholar]
- Palma, L.; Muñoz, D.; Berry, C.; Murillo, J.; Caballero, P. Bacillus thuringiensis toxins: An overview of their biocidal activity. Toxins (Basel) 2014, 6, 3296–3325. [Google Scholar] [CrossRef] [PubMed]
- Bravo, A.; Likitvivatanavong, S.; Gill, S.S.; Soberón, M. Bacillus thuringiensis: A story of a successful bioinsecticide. Insect Biochem. Mol. Biol. 2011, 41, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Crickmore, N.; Zeigler, D.R.; Feitelson, J.; Schnepf, E.; Van Rie, J.; Lereclus, D.; Baum, J.; Dean, D. Revision of the nomenclature for the Bacillus thuringiensis pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 1998, 62, 807–813. [Google Scholar] [PubMed]
- Schnepf, E.; Crickmore, N.; Van Rie, J.; Lereclus, D.; Baum, J.; Feitelson, J.; Zeigler, D.R.; Dean, D.H. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 1998, 62, 775–806. [Google Scholar] [PubMed]
- Vachon, V.; Laprade, R.; Schwartz, J.-L. Current models of the mode of action of Bacillus thuringiensis insecticidal crystal proteins: A critical review. J. Invertebr. Pathol. 2012, 111, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hofte, H.; Whiteley, H.R. Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol. Rev. 1989, 53, 242–255. [Google Scholar] [PubMed]
- El-Kersh, T.A.; Al-sheikh, Y.A.; Al-Akeel, R.; Alsayed, A.A. Isolation and characterization of indigenous Bacillus thuringiensis isolates from Saudi Arabia. Afr. J. Biotechnol. 2012, 11, 1924–1938. [Google Scholar]
- Ohba, M.; Mizuki, E.; Uemori, A. Parasporin, a new anticancer protein group from Bacillus thuringiensis. Anticancer Res. 2009, 29, 427–433. [Google Scholar] [PubMed]
- Mizuki, E.; Ohba, M.; Akao, T.; Yamashita, S.; Saitoh, H.; Park, Y.S. Unique activity associated with non-insecticidal Bacillus thuringiensis parasporal inclusions: In vitro cell-killing action on human cancer cells. J. Appl. Microbiol. 1999, 86, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Mizuki, E.; Park, Y.S.; Saitoh, H.; Yamashita, S.; Akao, T.; Higuchi, K.; Ohba, M. Parasporin, a human leukemic cell-recognizing parasporal protein of Bacillus thuringiensis. Clin. Diagn. Lab. Immunol. 2000, 7, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Katayama, H.; Yokota, H.; Akao, T.; Nakamura, O.; Ohba, M.; Mekada, E.; Mizuki, E. Parasporin-1, a novel cytotoxic protein to human cells from non-insecticidal parasporal inclusions of Bacillus thuringiensis. J. Biochem. 2005, 137, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, H.; Okumura, S.; Ishikawa, T.; Akao, T.; Mizuki, E.; Ohba, M. Investigation of a novel Bacillus thuringiensis gene encoding a parasporal protein, parasporin-4, that preferentially kills human cancer cells. Biosci. Biotechnol. Biochem. 2006, 70, 2935–2971. [Google Scholar] [CrossRef] [PubMed]
- Okumura, S.; Ohba, M.; Mizuki, E.; Crickmore, N.; Côté, J.-C.; Nagamatsu, Y.; Kitada, S.; Sakai, H.; Harata, K.; Shin, T. Parasporin Nomenclature. 2010. Available online: http://parasporin.fitc.pref.fukuoka.jp/ (accessed on 1 April 2013).
- Nagamatsu, Y.; Okamura, S.; Saitou, H.; Akao, T.; Mizuki, E. Three Cry toxins in two types from Bacillus thuringiensis strain M019 preferentially kill human hepatocyte cancer and uterus cervix cancer cells. Biosci. Biotechnol. Biochem. 2010, 74, 494–498. [Google Scholar] [CrossRef]
- Hire, R.S.; Makde, R.D.; Dongre, T.K.; D’Souza, S.F. Characterization of the cry1Ac17 gene from an indigenous strain of Bacillus thuringiensis subsp. kenyae. Curr. Microbiol. 2008, 57, 570–574. [Google Scholar] [CrossRef]
- Liang, H.; Liu, Y.; Zhu, J.; Guan, P.; Li, S.; Wang, S.; Zheng, A.; Liu, H.; Li, P. Characterization of cry2-type genes of Bacillus thuringiensis strains from soilisolated of Sichuan Basin, China. Braz. J. Microbiol. 2011, 42, 140–146. [Google Scholar] [CrossRef]
- Lakxmy, A.P.; Xavier, R.; Reenajosephine, C.M.; Lee, Y.W.; Marimuthu, K.; Kathiresan, S.; Sreeramanan, S. Mosquitocidal activity of a native Bacillus thuringiensis isolate Bt ReX02 from Gunung Jerai forest, Malaysia against Culex quinquefasciatus and Aedes albopictus. Eur. Rev. Med. Pharmacol. Sci. 2011, 15, 149–155. [Google Scholar]
- Lee, D.-W.; Akao, T.; Yamashita, S.; Katayama, H.; Maeda, M.; Saitoh, H.; Mizuki, E.; Ohba, M. Noninsecticidal parasporal proteins of a Bacillus thuringiensis serovar shandongiensis isolate exhibit a preferential cytotoxicity against human leukemic T cells. Biochem. Biophys. Res. Commun. 2000, 272, 218–223. [Google Scholar] [CrossRef]
- Nadarajah, V.D.; Ting, D.; Chan, K.K.; Mohamed, S.M.; Kanakeswary, K.; Lee, H.L. Selective cytotoxic activity against leukemic cell lines from mosquitocidal Bacillus thuringiensis parasporal inclusions. Southeast Asian J. Trop. Med. Public Health 2008, 39, 235–245. [Google Scholar] [PubMed]
- Poornima, K.; Saranya, V.; Abirami, P.; Binuramesh, C.; Suguna, P.; Selvanayagam, P.; Shenbagarathai, R. Phenotypic and genotypic characterization of B.t.LDC-391 strain that produce cytocidal proteins against human cancer cells. Bioinformation 2012, 8, 461–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chubicka, T.; Girija, D.; Deepa, K.; Salini, S.; Meera, N.; Raghavamenon, A.C.; Divya, M.K.; Babu, T.D. A parasporin from Bacillus thuringiensis native to Peninsular India induces apoptosis in cancer cells through intrinsic pathway. J. Biosci. 2018, 43, 407–416. [Google Scholar] [CrossRef] [PubMed]
- El-kersh, T.A.; Al-akeel, R.A.; Al-sheikh, Y.A.; Alharbi, S.A. Isolation and distribution of mosquito-larvicidal cry genes in Bacillus thuringiensis strains native to Saudi Arabia. Trop. Biomed. 2014, 31, 616–632. [Google Scholar] [PubMed]
- El-Kersh, T.A.; Ahmed, A.M.; Al-Sheikh, Y.A.; Tripet, F.; Ibrahim, M.S.; Metwalli, A.A. Isolation and characterization of native Bacillus thuringiensis strains from Saudi Arabia with enhanced larvicidal toxicity against the mosquito vector Anopheles gambiae (s.l.). Parasit Vectors 2016, 9, 647. [Google Scholar] [CrossRef] [PubMed]
- Svanborg, C.; Agerstam, H.; Aronson, A.; Bjerkvig, R.; Duringer, C.; Fischer, W.; Gustafsson, L.; Hallgren, O.; Leijonhuvud, I.; Linse, S.; et al. HAMLET kills tumor cells by an apoptosis-like mechanism-cellular, molecular, and therapeutic aspects. Adv. Cancer Res. 2003, 88, 1–29. [Google Scholar] [PubMed]
- Jia, L.-T.; Chen, S.Y.; Yang, A.-G. Cancer gene therapy targeting cellular apoptosis machinery. Cancer Treat. Rev. 2012, 38, 868–876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruden, M.; Puri, N. Novel anticancer therapeutics targeting telomerase. Cancer Treat. Rev. 2013, 39, 444–456. [Google Scholar] [CrossRef]
- Chen, C.-C.; Chang, F.-H.; Lin, K.-H.; Tsai, D.-H.; Chen, Y.-Y. Photothermal cancer therapy via femtosecond-laser-excited FePt nanoparticles. Biomaterials 2013, 34, 1128–1134. [Google Scholar] [CrossRef]
- Kumar, A.; Kant, S.; Singh, S.M. Novel molecular mechanisms of antitumor action of dichloroacetate against T cell lymphoma: Implication of altered glucose metabolism, pH homeostasis and cell survival regulation. Chem. Biol. Interact. 2012, 199, 29–37. [Google Scholar] [CrossRef]
- Glazer, E.S.; Massey, K.L.; Zhu, C.; Curley, S.A. Pancreatic carcinoma cells are susceptible to noninvasive radio frequency fields after treatment with targeted gold nanoparticles. Surgery 2010, 148, 319–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben-Arye, E.; Lev, E.; Schiff, E. Complementary Medicine Oncology Research in the Middle-East: Shifting from Traditional to Integrative Cancer Care. Eur. J. Integr. Med. 2011, 3, 29–37. [Google Scholar] [CrossRef]
- Hussain, S.S.; Kumar, A.P.; Ghosh, R. Food-based natural products for cancer management: Is the whole greater than the sum of the parts? Semin. Cancer Biol. 2016, 40–41, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Patyar, S.; Joshi, R.; Prasad Byrav, D.S.; Prakash, A.; Medhi, B.; Das, B.K. Bacteria in cancer therapy: A novel experimental strategy. J. Biomed. Sci. 2010, 17, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.M.; Hussein, H.I.; El-Kersh, T.A.; Al-Sheikh, Y.A.; Ayaad, T.H.; El-Sadawy, H.A.; Al-Mekhlafi, F.A.; Ibrahim, M.S.; Al-Tamimi, J.; Nasr, F.A. Larvicidal activities of indigenous Bacillus thuringiensis isolates and nematode symbiotic bacterial toxins against the mosquito vector, Culex pipiens (Diptera: Culicidae). J. Arthropod-Borne Dis. 2017, 11, 260–277. [Google Scholar] [PubMed]
- Martin, P.A.; Gundersen-Rindal, D.E.; Blackburn, M.B. Distribution of phenotypes among Bacillus thuringiensis strains. Syst. Appl. Microbiol. 2010, 33, 204–208. [Google Scholar] [CrossRef] [PubMed]
- Ito, A.; Sasaguri, Y.; Kitada, S.; Kusakar, Y.; Kuwano, K.; Masutomi, K.; Mizuki, E.; Akao, T.; Ohba, M. A Bacillus thuringiensis crystal protein with selective cytocidal action to human cells. J. Biol. Chem. 2004, 279, 21282–21286. [Google Scholar] [CrossRef] [PubMed]
- Ibarra, J.E.; del Rincon, M.C.; Orduz, S.; Noriega, D.; Benintende, G.; Monnerat, R.; Regis, L.; de Oliveira, C.M.; Lanz, H.; Rodriguez, M.H.; et al. Diversity of Bacillus thuringiensis strains from Latin America with insecticidal activity against different mosquito species. Appl. Environ. Microbiol. 2003, 69, 5269–5274. [Google Scholar] [CrossRef] [PubMed]
- Akiba, T.; Okumura, S. Parasporins 1 and 2: Their structure and activity. J. Invertebr. Pathol. 2017, 142, 44–49. [Google Scholar] [CrossRef]
- de Maagd, R.A.; Bravo, A.; Berry, C.; Crickmore, N.; Schnepf, H.E. Structure, diversity, and evolution of protein toxins from spore-forming entomopathogenic bacteria. Annu. Rev. Genet. 2003, 37, 409–433. [Google Scholar] [CrossRef]
- Yamashita, S.; Katayama, H.; Saitoh, H.; Akao, T.; Park, Y.S.; Mizuki, E.; Ohba, M.; Ito, A. Typical Three-Domain Cry Proteins of Bacillus thuringiensis Strain A1462 Exhibit Cytocidal Activity on Limited Human Cancer Cells. J. Biochem. 2005, 138, 663–672. [Google Scholar] [CrossRef]
- Katayama, H.; Kusaka, Y.; Yokota, H.; Akao, T.; Kojima, M.; Nakamura, O.; Mekada, E.; Mizuki, E. Parasporin-1, a novel cytotoxic protein from Bacillus thuringiensis, induces Ca2+ influx and a sustained elevation of the cytoplasmic Ca2+ concentration in toxin-sensitive cells. J. Biol. Chem. 2007, 282, 7742–7752. [Google Scholar] [CrossRef] [PubMed]
- Green, D.R.; Amarante-Mendes, G.P. The point-of-no-return: Mitochondria, caspases and the commitment to cell death. Results Probl. Cell Differ. 1998, 24, 45–61. [Google Scholar] [PubMed]
- Sanger, F.; Nicklem, S.; Coulso, N.A. DNA sequencing using chain termination inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [PubMed]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. 1999, 41, 95–98. [Google Scholar]
- Yu, Y.M.; Ohba, M.; Gill, S.S. Characterization of mosquitocidal activity of Bacillus thuringiensis subsp fukuokaensis crystal proteins. Appl. Environ. Microbiol. 1991, 57, 1075–1081. [Google Scholar] [PubMed]
- Nethravathi, C.J.; Hugar, P.S.; Krishnaraj, P.U.; Vastrad, A.S.; Awaknavar, J.S. Bioefficacy of crude protein of native Bacillus thuringiensis (Berliner) isolates against cabbage leaf. Karnataka J. Agric. Sci. 2009, 22, 613–616. [Google Scholar]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Blum, H.; Beier, H.; Gross, H.J. Improved method for silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 1987, 8, 93–99. [Google Scholar] [CrossRef]
- Kohle, C.; Badary, O.A.; Nill, K.; Bock-Hennig, B.S.; Bock, K.W. Serotonin glucuronidation by Ah receptor- and oxidative stress-inducible human UDP-glucuronosyltransferase (UGT) 1A6 in Caco-2 cells. Biochem. Pharmacol. 2005, 69, 1397–1402. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, L.J.; Lee, F.Y.; Chen, P.; Norris, D.; Barrish, J.C.; Behnia, K.; Castaneda, S.; Cornelius, L.A.; Das, J.; Doweyko, A.; et al. Discovery of N-(2-chloro-6-methyl-phenyl)-2-(6-(4-(2-hydroxyethyl)-piperazin-1-yl)-2-methylpyrimidin-4-ylamino)thiazole-5-carboxamide (BMS-354825), a dual Src/Abl kinase inhibitor with potent antitumor activity in preclinical assays. J. Med. Chem. 2004, 47, 6658–6661. [Google Scholar] [CrossRef] [PubMed]
- Al-Sheikh, Y.A.; Ghneim, H.K.; Softa, K.I.; Al-Jobran, A.A.; Al-Obeed, O.; Mohamed, M.A.; Abdulla, M.; Aboul-Soud, M.A. Expression profiling of selected microRNA signatures in plasma and tissues of Saudi colorectal cancer patients by qPCR. Oncol. Lett. 2016, 11, 1406–1412. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |




| Isolate/Serial# | Geographical Location/Crystal Shape | SAC | AMY | VP | CIT | OX | MOT | ADH |
|---|---|---|---|---|---|---|---|---|
| Bt1 (284) | Al Hasa/Bi-pyramidal not attached | + | + | + | + | + | + | + |
| Bt2 (319) | Hafr Al-Baten/Attached hexagonal | − | + | + | + | + | − | + |
| Bt3 (594) | Yonbaa/Spherical | − | + | − | + | − | + | − |
| Bt4 (605) | Al-Majma’ah/Spherical | + | + | − | − | + | + | + |
| Bt5 (M300) | Mecca/Dark, bright, ovoid, Attached spore | + | + | + | + | + | + | + |
| Bt6 (M268) | Al-Madina/Hexagonal-spore in chain | − | − | + | − | + | − | − |
| Bt7 (M13m) | Asir/Small, outside, spherical, not attached | − | + | − | + | + | − | + |
| Bt8 (M160) | Al-Majma’ah/Large bi-pyramidal, attached spore | − | − | + | − | + | − | − |
| Bt9 (224) | Al Taif/Hexagonal, inclined spore | − | + | + | + | + | − | + |
| Bt isolate/Compound | HeLa | HT-29 | ||
|---|---|---|---|---|
| Inhibition (%) * | IC50 # (µg/mL) | Inhibition * (%) | IC50 (µg/mL) | |
| Bt1 | 38 | 3.2 | 3 | nd a |
| Bt2 | 40 | 5.7 | 2 | nd |
| Bt3 | 35 | 5.4 | 0 | nd |
| Bt4 | 38 | 5.4 | 2 | nd |
| Bt5 | 40 | 9.5 | 3 | nd |
| Bt6 | 42 | 14.2 | 0 | nd |
| Bt7 | 47 | 5.6 | 0 | nd |
| Bt8 | 40 | 6.4 | 0 | nd |
| Bt9 | 43 | 6.1 | 4 | nd |
| Dasatinib | 58 | 24.4 | nd | nd |
| Primer | Sequence | Gene(s) Recognized | Amplicon Size (bP) | Condition of Annealing | GenBank Accession No | Reference |
|---|---|---|---|---|---|---|
| Cry2 (UN) | (F)5′-GAGTTTAATCGACAAGTAGATAATTT-3′ (R)5′-GGAAAAGAGAATATAAAAATGGCCAG-3′ | Cry2Aa | 526 | 50 °C | M31738 | [41] |
| Cry2Ab | 526 | M23724 | ||||
| Cry2Ac | 520 | X57252 | ||||
| Cry2Ad | 500 | AF200816 | ||||
| Cry4 (UN) | (F)5′-GCATATGATGTAGCGAAACAAGCC-3′ (R)5′-GCGTGACATACCCATTTCCAGGTCC-3′ | Cry4A2 | 439 | 58 °C | D00248 | [27] |
| Cry4B4 | 439 | D00247 | ||||
| Cry4A (spe) | (F)5′-TCAAAGATCATTTCAAAATTACATG-3′ (R)5′-CGGCTTGATCTATGTCATAATCTGT-3′ | Cry4Aa | 459 | 50 °C | Y00423 | [27] |
| Cry4B (spe) | (F)5′-CGTTTCAAGACCTAATAATATAATACC-3′ (R)5′-CGGCTTGATCTATGTCATAATCTGT-3′ | Cry4Ba | 321 | 50 °C | ‘X07423 | [27] |
| Cry10 (spe) | (F)5’-TCAATGCTCCATCCAATG-3′ (R)5’-CTTGTATAGGCCTTCCTCCG-3′ | Cry10 | 348 | 51 °C | M12662 | [27] |
| Cry11 (UN) | (F)5’-CGCTTACAGGATGGATAGG-3′ (R)5′-GCTGAAACGGCACGAATATAATA-3′ | Cry11Aa | 342 | 50 °C | M31737 | [41] |
| Cry11Ba | 342 | X86902 | ||||
| Cry11Bb | 452 | AF017416 | ||||
| Cyt1 (UN) | (F)5′-CCTCAATCAACAGCAAGGGTTATT-3′ (R)5′-TGCAAACAGGACATTGTATGTGTAATT-3′ | Cyt1Aa | 477 | 52 °C | X03182 | [41] |
| Cyt1Ab | 480 | X98793 | ||||
| Cyt1Ba | 477 | U37196 | ||||
| Cyt2 (UN) | (F)5′-ATTACAAATTGCAAATGGTATTCC-3′ (R)5′-TTTCAACATCCACAGTAATTTCAAATGC-3′ | Cyt2Aa | 356 | 50 °C | Z14147 | [41] |
| Cyt2Ba | 355 | U52043 | ||||
| Cyt2Bb | 355 | U82519 | ||||
| Cyt2Ca | 355 | AAK50455 | ||||
| PS1Aa1 | (F)5′-TGTGCGATTGGTGGATGCGCT-3′ (R)5′-TCCCCGAAAAAGACCTGCGGT-3′ | Cry31Aa1 | 226 | 65 °C | AB031065 | [15] |
| PS2Aa1 | (F)5′-TCCCAAAAGAGTAGGGCCAGGTG-3′ (R)3′-AATTCCCCCATTTTGGGCATTGGCA-3′ | Cry46Aa1 | 269 | 63 °C | AB099515 | [41] |
| PS3Aa1 | (F)5′-GCCGGAATTGCTGGCCTCGA-3′ (R)5′-TGATGGGCTCCGTAGGTAGGGA-3′ | Cry41Aa1 | 447 | 52 °C | AB116649 | [4] |
| PS4Aa1 | (F)5′-GAGGTGGTGTGCTGCAAGGGG-3′ (R)5′-TTCCCGAACCTGCCCTGCAC-3′ | Cry45Aa1 | 712 | 57 °C | AB180980 | [17] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aboul-Soud, M.A.M.; Al-Amri, M.Z.; Kumar, A.; Al-Sheikh, Y.A.; Ashour, A.E.; El-Kersh, T.A. Specific Cytotoxic Effects of Parasporal Crystal Proteins Isolated from Native Saudi Arabian Bacillus thuringiensis Strains against Cervical Cancer Cells. Molecules 2019, 24, 506. https://doi.org/10.3390/molecules24030506
Aboul-Soud MAM, Al-Amri MZ, Kumar A, Al-Sheikh YA, Ashour AE, El-Kersh TA. Specific Cytotoxic Effects of Parasporal Crystal Proteins Isolated from Native Saudi Arabian Bacillus thuringiensis Strains against Cervical Cancer Cells. Molecules. 2019; 24(3):506. https://doi.org/10.3390/molecules24030506
Chicago/Turabian StyleAboul-Soud, Mourad A. M., Mohammed Z. Al-Amri, Ashok Kumar, Yazeed A. Al-Sheikh, Abdelkader E. Ashour, and Talat A. El-Kersh. 2019. "Specific Cytotoxic Effects of Parasporal Crystal Proteins Isolated from Native Saudi Arabian Bacillus thuringiensis Strains against Cervical Cancer Cells" Molecules 24, no. 3: 506. https://doi.org/10.3390/molecules24030506
APA StyleAboul-Soud, M. A. M., Al-Amri, M. Z., Kumar, A., Al-Sheikh, Y. A., Ashour, A. E., & El-Kersh, T. A. (2019). Specific Cytotoxic Effects of Parasporal Crystal Proteins Isolated from Native Saudi Arabian Bacillus thuringiensis Strains against Cervical Cancer Cells. Molecules, 24(3), 506. https://doi.org/10.3390/molecules24030506








