Molecular Pathological Diagnosis of Thyroid Tumors Using Spatially Resolved Metabolomics
Abstract
:1. Introduction
2. Results
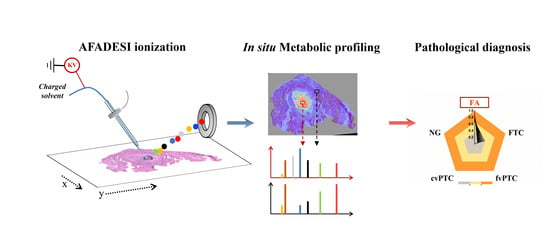
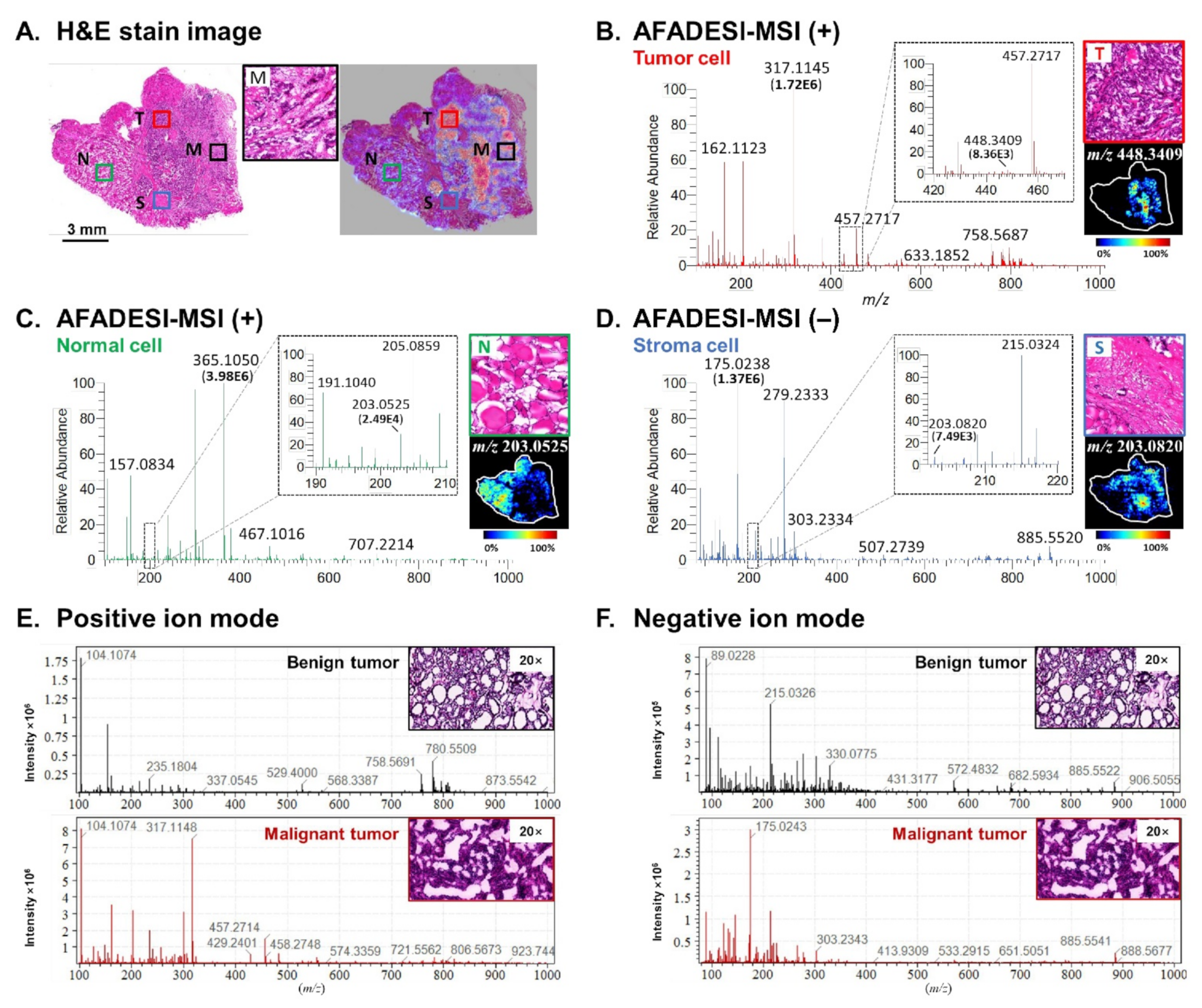
2.1. Spatially Resolved Metabolic Profiling
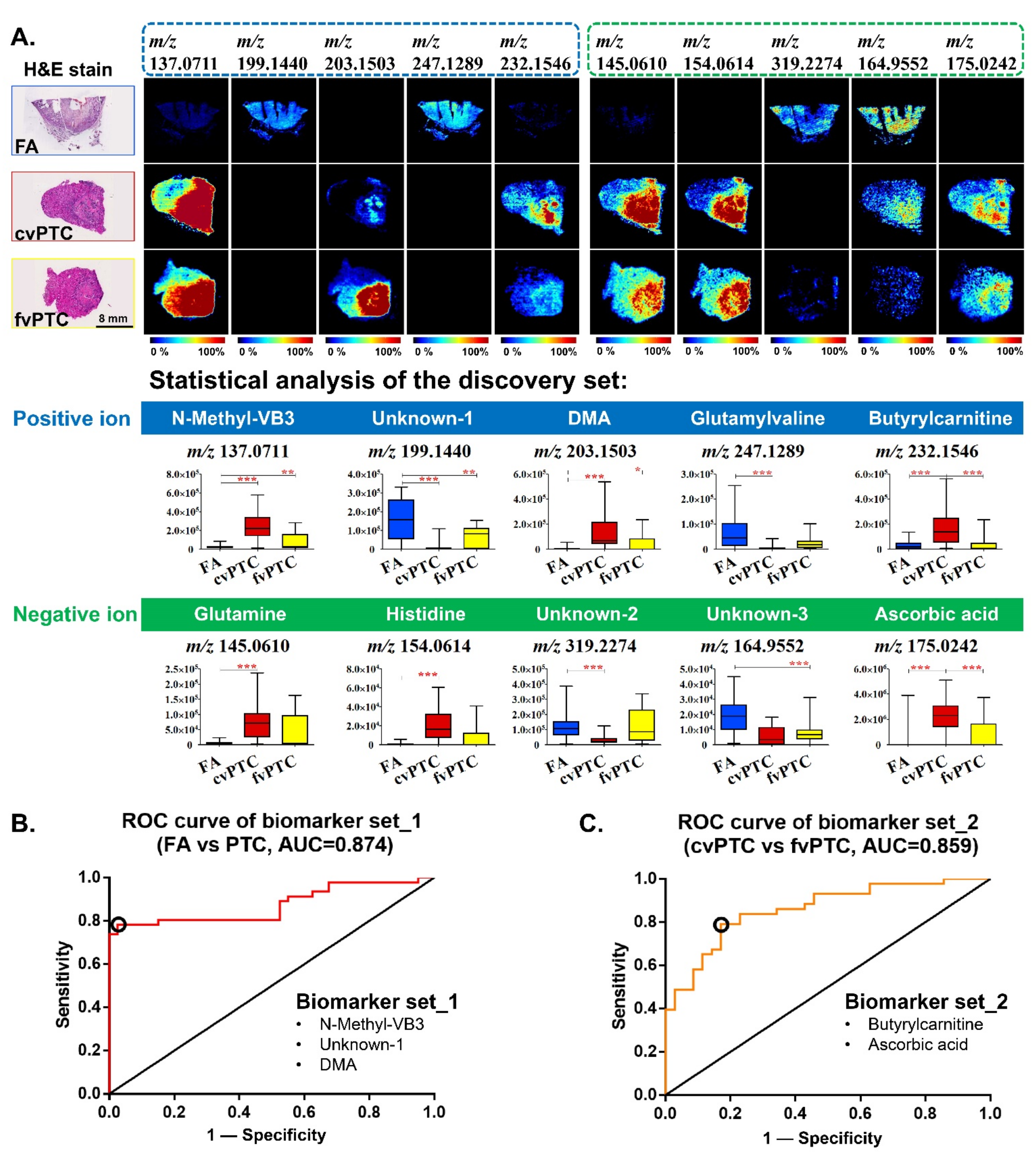
2.2. Biomarkers for Discriminating Benign FA from Malignant PTC
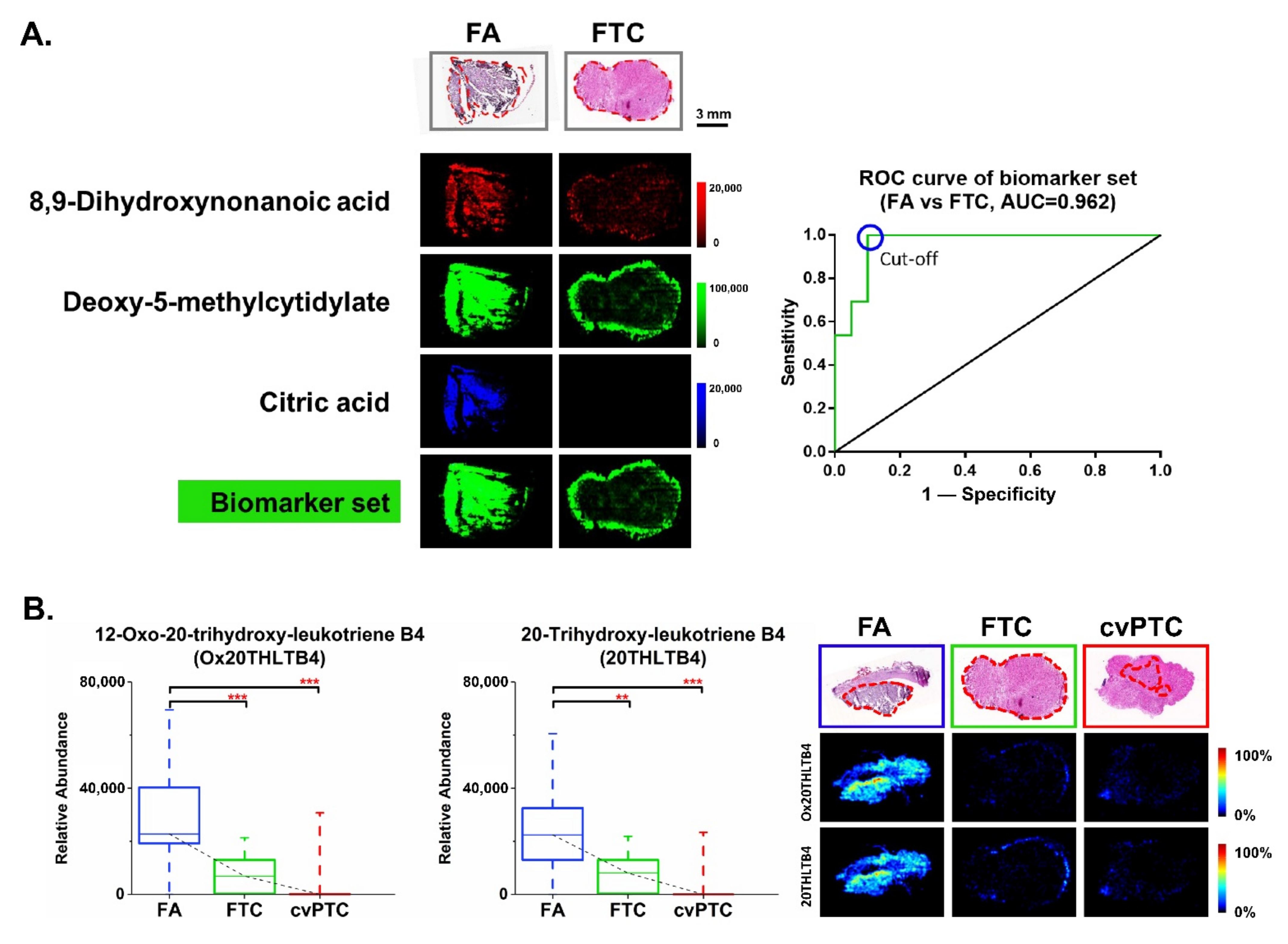
2.3. Biomarkers for Discriminating Benign FA from Malignant FTC
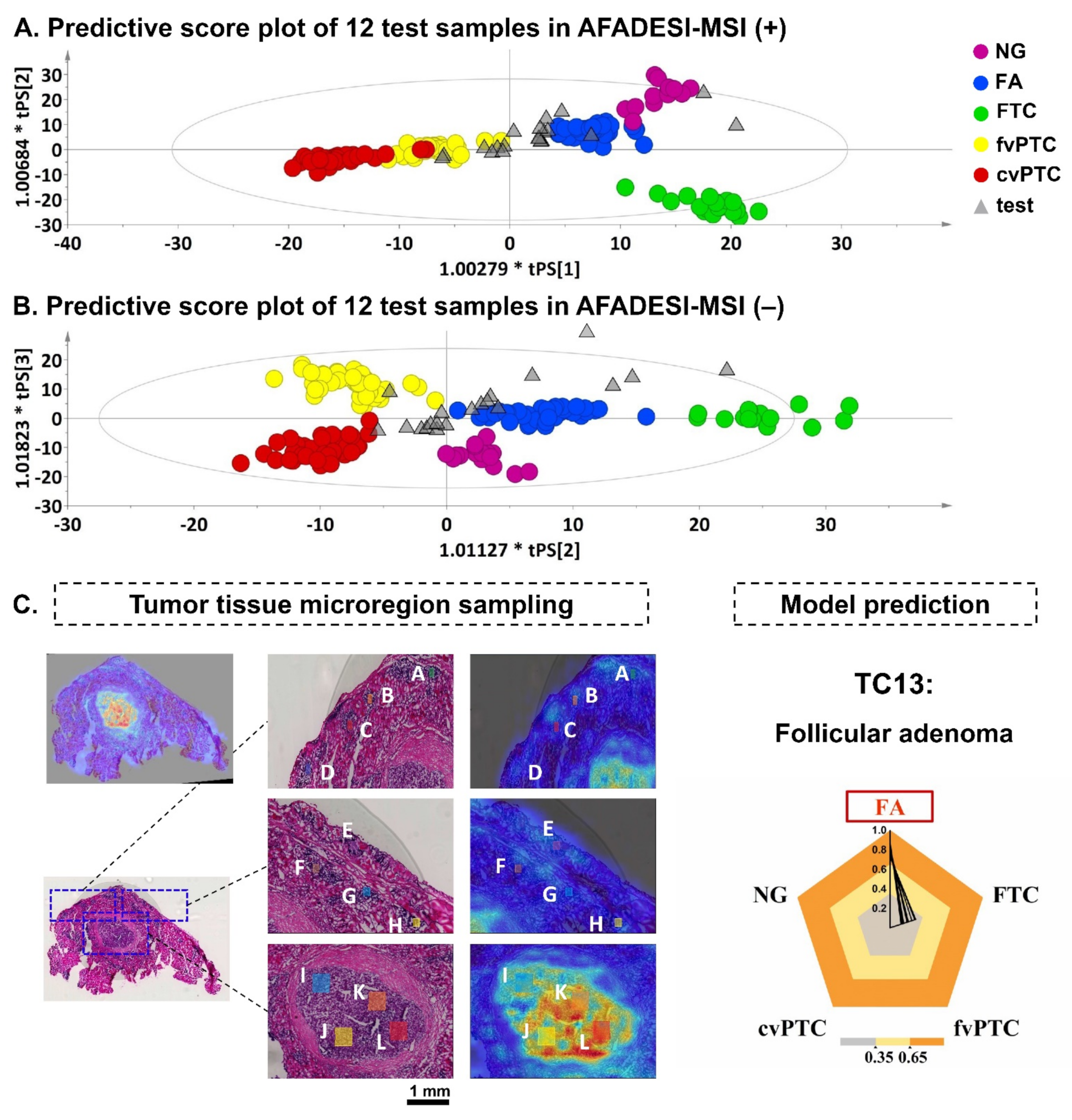
2.4. Diagnostic Model and Discriminant Analysis
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Preparation
4.2. AFADESI-MSI Experiment
4.3. Data Processing
4.4. Metabolite Identification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodarzi, E.; Moslem, A.; Feizhadad, H.; Jarrahi, A.; Adineh, H.; Sohrabivafa, M.; Khazaei, Z. Epidemiology, incidence and mortality of thyroid cancer and their relationship with the human development index in the world: An ecology study in 2018. Adv. Hum. Biol. 2019, 9, 162–167. [Google Scholar]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting Cancer Incidence and Deaths to 2030: The Unexpected Burden of Thyroid, Liver, and Pancreas Cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [Green Version]
- Poller, D.N.; Glaysher, S. Molecular pathology and thyroid FNA. Cytopathology 2017, 28, 475–481. [Google Scholar] [CrossRef]
- Baloch, Z.; LiVolsi, V.A. Fifty years of thyroid pathology: Concepts and developments. Hum. Pathol. 2020, 95, 46–54. [Google Scholar] [CrossRef]
- Vasko, V.V.; Gaudart, J.; Allasia, C.; Savchenko, V.; Di Cristofaro, J.; Saji, M.; Ringel, M.D.; De Micco, C. Thyroid follicular adenomas may display features of follicular carcinoma and follicular variant of papillary carcinoma. Eur. J. Endocrinol. 2004, 151, 779–786. [Google Scholar] [CrossRef] [Green Version]
- Yoon, J.H.; Kim, E.-K.; Youk, J.H.; Moon, H.J.; Kwak, J.Y. Better understanding in the differentiation of thyroid follicular adenoma, follicular carcinoma, and follicular variant of papillary carcinoma: A retrospective study. Int. J. Endocrinol. 2014, 2014, 321595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sreekumar, A.; Poisson, L.M.; Rajendiran, T.M.; Khan, A.P.; Cao, Q.; Yu, J.; Laxman, B.; Mehra, R.; Lonigro, R.J.; Li, Y.; et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 2009, 457, 910–914. [Google Scholar] [CrossRef] [Green Version]
- Newgard, C.B. Metabolomics and Metabolic Diseases: Where Do We Stand? Cell Metab. 2017, 25, 43–56. [Google Scholar] [CrossRef] [Green Version]
- Levine, A.J.; Puzio-Kuter, A.M. The Control of the Metabolic Switch in Cancers by Oncogenes and Tumor Suppressor Genes. Science 2010, 330, 1340. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Zheng, Y.; Zhao, B.; Zhang, Y.; Liu, Z.; Xu, J.; Chen, Y.; Yang, Z.; Wang, F.; Wang, H.; et al. Human Metabolic Responses to Chronic Environmental Polycyclic Aromatic Hydrocarbon Exposure by a Metabolomic Approach. J. Proteome Res. 2015, 14, 2583–2593. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.L.; Liu, D.; Ho, M.-F.; Athreya, A.P.; Weinshilboum, R. Selective Serotonin Reuptake Inhibitor Pharmaco-Omics: Mechanisms and Prediction. Front. Pharmacol. 2021, 11, 2278. [Google Scholar] [CrossRef] [PubMed]
- Rubakhin, S.S.; Jurchen, J.C.; Monroe, E.B.; Sweedler, J.V. Imaging mass spectrometry: Fundamentals and applications to drug discovery. Drug Discov. Today 2005, 10, 823–837. [Google Scholar] [CrossRef]
- Porta Siegel, T.; Hamm, G.; Bunch, J.; Cappell, J.; Fletcher, J.S.; Schwamborn, K. Mass Spectrometry Imaging and Integration with Other Imaging Modalities for Greater Molecular Understanding of Biological Tissues. Mol. Imaging Biol. 2018, 20, 888–901. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Johnson, J.; Wang, B.; Chen, B.; Fisher, G.L.; Urabe, G.; Shi, X.; Kent, K.C.; Guo, L.-W.; Li, L. Mass Spectrometric Imaging Reveals Temporal and Spatial Dynamics of Bioactive Lipids in Arteries Undergoing Restenosis. J. Proteome Res. 2019, 18, 1669–1678. [Google Scholar] [CrossRef]
- Xie, P.; Liang, X.; Song, Y.; Cai, Z. Mass Spectrometry Imaging Combined with Metabolomics Revealing the Proliferative Effect of Environmental Pollutants on Multicellular Tumor Spheroids. Anal. Chem. 2020, 92, 11341–11348. [Google Scholar] [CrossRef]
- Cooks, R.G.; Ouyang, Z.; Takats, Z.; Wiseman, J.M. Ambient Mass Spectrometry. Science 2006, 311, 1566–1570. [Google Scholar] [CrossRef]
- Harris, G.A.; Galhena, A.S.; Fernández, F.M. Ambient Sampling/Ionization Mass Spectrometry: Applications and Current Trends. Anal. Chem. 2011, 83, 4508–4538. [Google Scholar] [CrossRef]
- Zhang, J.; Yu, W.; Ryu, S.W.; Lin, J.; Buentello, G.; Tibshirani, R.; Suliburk, J.; Eberlin, L.S. Cardiolipins Are Biomarkers of Mitochondria-Rich Thyroid Oncocytic Tumors. Cancer Res. 2016, 76, 6588–6597. [Google Scholar] [CrossRef] [Green Version]
- Porcari, A.M.; Zhang, J.; Garza, K.Y.; Rodrigues-Peres, R.M.; Lin, J.Q.; Young, J.H.; Tibshirani, R.; Nagi, C.; Paiva, G.R.; Carter, S.A.; et al. Multicenter Study Using Desorption-Electrospray-Ionization-Mass-Spectrometry Imaging for Breast-Cancer Diagnosis. Anal. Chem. 2018, 90, 11324–11332. [Google Scholar] [CrossRef]
- Margulis, K.; Chiou, A.S.; Aasi, S.Z.; Tibshirani, R.J.; Tang, J.Y.; Zare, R.N. Distinguishing malignant from benign microscopic skin lesions using desorption electrospray ionization mass spectrometry imaging. Proc. Natl. Acad. Sci. USA 2018, 115, 6347–6352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, J.; Sun, C.; Li, T.; Luo, Z.; Huang, L.; Song, X.; Li, X.; Abliz, Z. A Sensitive and Wide Coverage Ambient Mass Spectrometry Imaging Method for Functional Metabolites Based Molecular Histology. Adv. Sci. 2018, 5, 1800250. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Li, T.; Song, X.; Huang, L.; Zang, Q.; Xu, J.; Bi, N.; Jiao, G.; Hao, Y.; Chen, Y.; et al. Spatially resolved metabolomics to discover tumor-associated metabolic alterations. Proc. Natl. Acad. Sci. USA 2019, 116, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Min, K.-W.; Bang, J.-Y.; Kim, K.P.; Kim, W.-S.; Lee, S.H.; Shanta, S.R.; Lee, J.H.; Hong, J.H.; Lim, S.D.; Yoo, Y.-B.; et al. Imaging mass spectrometry in papillary thyroid carcinoma for the identification and validation of biomarker proteins. J. Korean Med. Sci. 2014, 29, 934–940. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, S.; Tateya, I.; Hayasaka, T.; Masaki, N.; Takizawa, Y.; Ohno, S.; Kojima, T.; Kitani, Y.; Kitamura, M.; Hirano, S.; et al. Increased Expression of Phosphatidylcholine (16:0/18:1) and (16:0/18:2) in Thyroid Papillary Cancer. PLoS ONE 2012, 7, e48873. [Google Scholar] [CrossRef] [Green Version]
- Galli, M.; Pagni, F.; De Sio, G.; Smith, A.; Chinello, C.; Stella, M.; L'Imperio, V.; Manzoni, M.; Garancini, M.; Massimini, D.; et al. Proteomic profiles of thyroid tumors by mass spectrometry-imaging on tissue microarrays. Biochim. Biophys. Acta 2017, 1865, 817–827. [Google Scholar] [CrossRef]
- Pagni, F.; De Sio, G.; Garancini, M.; Scardilli, M.; Chinello, C.; Smith, A.J.; Bono, F.; Leni, D.; Magni, F. Proteomics in thyroid cytopathology: Relevance of MALDI-imaging in distinguishing malignant from benign lesions. Proteomics 2016, 16, 1775–1784. [Google Scholar] [CrossRef]
- Pietrowska, M.; Diehl, H.C.; Mrukwa, G.; Kalinowska-Herok, M.; Gawin, M.; Chekan, M.; Elm, J.; Drazek, G.; Krawczyk, A.; Lange, D.; et al. Molecular profiles of thyroid cancer subtypes: Classification based on features of tissue revealed by mass spectrometry imaging. Biochim. Biophys. Acta 2017, 1865, 837–845. [Google Scholar] [CrossRef] [Green Version]
- Nipp, M.; Elsner, M.; Balluff, B.; Meding, S.; Sarioglu, H.; Ueffing, M.; Rauser, S.; Unger, K.; Höfler, H.; Walch, A.; et al. S100-A10, thioredoxin, and S100-A6 as biomarkers of papillary thyroid carcinoma with lymph node metastasis identified by MALDI imaging. J. Mol. Med. 2012, 90, 163–174. [Google Scholar] [CrossRef]
- DeHoog, R.J.; Zhang, J.; Alore, E.; Lin, J.Q.; Yu, W.; Woody, S.; Almendariz, C.; Lin, M.; Engelsman, A.F.; Sidhu, S.B.; et al. Preoperative metabolic classification of thyroid nodules using mass spectrometry imaging of fine-needle aspiration biopsies. Proc. Natl. Acad. Sci. USA 2019, 116, 21401–21408. [Google Scholar] [CrossRef] [Green Version]
- Xuan, J.; Pan, G.; Qiu, Y.; Yang, L.; Su, M.; Liu, Y.; Chen, J.; Feng, G.; Fang, Y.; Jia, W.; et al. Metabolomic Profiling to Identify Potential Serum Biomarkers for Schizophrenia and Risperidone Action. J. Proteome Res. 2011, 10, 5433–5443. [Google Scholar] [CrossRef]
- Murphy, R.C.; Gijón, M.A. Biosynthesis and metabolism of leukotrienes. Biochem. J. 2007, 405, 379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Archambault, A.-S.; Poirier, S.; Lefebvre, J.-S.; Robichaud, P.-P.; Larose, M.-C.; Turcotte, C.; Martin, C.; Provost, V.; Boudreau, L.H.; McDonald, P.P.; et al. 20-Hydroxy- and 20-carboxy-leukotriene (LT) B4 downregulate LTB4-mediated responses of human neutrophils and eosinophils. J. Leukoc. Biol. 2019, 105, 1131–1142. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [Green Version]
- Cairns, R.A.; Harris, I.S.; Mak, T.W. Regulation of cancer cell metabolism. Nat. Rev. Cancer 2011, 11, 85. [Google Scholar] [CrossRef] [Green Version]
- Teng, H.; Mao, F.; Liang, J.; Xue, M.; Wei, W.; Li, X.; Zhang, K.; Feng, D.; Liu, B.; Sun, Z. Transcriptomic signature associated with carcinogenesis and aggressiveness of papillary thyroid carcinoma. Theranostics 2018, 8, 4345–4358. [Google Scholar] [CrossRef]
- Hanigan, M.H. Gamma-glutamyl transpeptidase: Redox regulation and drug resistance. Adv. Cancer Res. 2014, 122, 103–141. [Google Scholar] [PubMed] [Green Version]
- Noonan, D.M.; De Lerma Barbaro, A.; Vannini, N.; Mortara, L.; Albini, A. Inflammation, inflammatory cells and angiogenesis: Decisions and indecisions. Cancer Metastasis Rev. 2008, 27, 31–40. [Google Scholar] [CrossRef]
- Wang, D.; Dubois, R.N. Eicosanoids and cancer. Nat. Rev. Cancer 2010, 10, 181–193. [Google Scholar] [CrossRef]
- Luo, Z.; He, J.; Chen, Y.; He, J.; Gong, T.; Tang, F.; Wang, X.; Zhang, R.; Huang, L.; Zhang, L.; et al. Air Flow-Assisted Ionization Imaging Mass Spectrometry Method for Easy Whole-Body Molecular Imaging under Ambient Conditions. Anal. Chem. 2013, 85, 2977–2982. [Google Scholar] [CrossRef]
- Lv, Y.; Li, T.; Guo, C.; Sun, C.; Tang, F.; Huang, L.; Luo, Z.; Li, X.; Zhang, R.; Zang, Q.; et al. A high-performance bio-tissue imaging method using air flow-assisted desorption electrospray ionization coupled with a high-resolution mass spectrometer. Chin. Chem. Lett. 2019, 30, 461–464. [Google Scholar] [CrossRef]
- He, J.; Huang, L.; Tian, R.; Li, T.; Sun, C.; Song, X.; Lv, Y.; Luo, Z.; Li, X.; Abliz, Z. MassImager: A software for interactive and in-depth analysis of mass spectrometry imaging data. Anal. Chim. Acta 2018, 1015, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, R.A.; Hoefsloot, H.C.J.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [Green Version]
- Wojakowska, A.; Chekan, M.; Widlak, P.; Pietrowska, M. Application of Metabolomics in Thyroid Cancer Research. Int. J. Endocrinol. 2015, 2015, 258763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, S.; Qiu, L.; Wang, Y.; Qin, X.; Liu, H.; He, M.; Zhang, Y.; Li, Z.; Chen, X. Tissue imaging and serum lipidomic profiling for screening potential biomarkers of thyroid tumors by matrix-assisted laser desorption/ionization-Fourier transform ion cyclotron resonance mass spectrometry. Anal. Bioanal. Chem. 2014, 406, 4357–4370. [Google Scholar] [CrossRef]
- Zhang, J.; Feider, C.L.; Nagi, C.; Yu, W.; Carter, S.A.; Suliburk, J.; Cao, H.S.T.; Eberlin, L.S. Detection of Metastatic Breast and Thyroid Cancer in Lymph Nodes by Desorption Electrospray Ionization Mass Spectrometry Imaging. J. Am. Soc. Mass Spectrom. 2017, 28, 1166–1174. [Google Scholar] [CrossRef] [PubMed]
| Positive Mode | Negative Mode | Computational Prediction | ||||
|---|---|---|---|---|---|---|
| Class FA | Class FTC | Class FA | Class FTC | Class FA | Class FTC | |
| TC13-P-1-A | 0.9262 | 0.0738 | 0.6859 | 0.3141 | 0.8060 | 0.1940 |
| TC13-P-1-B | 0.9632 | 0.0368 | 0.7837 | 0.2163 | 0.8735 | 0.1265 |
| TC13-P-1-C | 0.9195 | 0.0805 | 0.8438 | 0.1562 | 0.8817 | 0.1183 |
| TC13-P-1-D | 0.8737 | 0.1263 | 0.7340 | 0.2660 | 0.8038 | 0.1962 |
| TC13-P-2-E | 0.9160 | 0.0840 | 0.5535 | 0.4465 | 0.7347 | 0.2653 |
| TC13-P-2-F | 0.7397 | 0.2603 | 0.7103 | 0.2897 | 0.7250 | 0.2750 |
| TC13-P-2-G | 0.9898 | 0.0102 | 0.7180 | 0.2820 | 0.8539 | 0.1461 |
| TC13-P-2-H | 1.5810 | −0.5810 | 0.6761 | 0.3239 | 1.1286 | −0.1286 |
| TC13-P-3-I | 0.8098 | 0.1902 | 0.7215 | 0.2785 | 0.7656 | 0.2344 |
| TC13-P-3-J | 0.8937 | 0.1063 | 0.8215 | 0.1785 | 0.8576 | 0.1424 |
| TC13-P-3-K | 0.9020 | 0.0980 | 0.8260 | 0.1740 | 0.8640 | 0.1360 |
| TC13-P-3-L | 0.8442 | 0.1558 | 0.7925 | 0.2075 | 0.8183 | 0.1817 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, L.; Mao, X.; Sun, C.; Li, T.; Song, X.; Li, J.; Gao, S.; Zhang, R.; Chen, J.; He, J.; et al. Molecular Pathological Diagnosis of Thyroid Tumors Using Spatially Resolved Metabolomics. Molecules 2022, 27, 1390. https://doi.org/10.3390/molecules27041390
Huang L, Mao X, Sun C, Li T, Song X, Li J, Gao S, Zhang R, Chen J, He J, et al. Molecular Pathological Diagnosis of Thyroid Tumors Using Spatially Resolved Metabolomics. Molecules. 2022; 27(4):1390. https://doi.org/10.3390/molecules27041390
Chicago/Turabian StyleHuang, Luojiao, Xinxin Mao, Chenglong Sun, Tiegang Li, Xiaowei Song, Jiangshuo Li, Shanshan Gao, Ruiping Zhang, Jie Chen, Jiuming He, and et al. 2022. "Molecular Pathological Diagnosis of Thyroid Tumors Using Spatially Resolved Metabolomics" Molecules 27, no. 4: 1390. https://doi.org/10.3390/molecules27041390