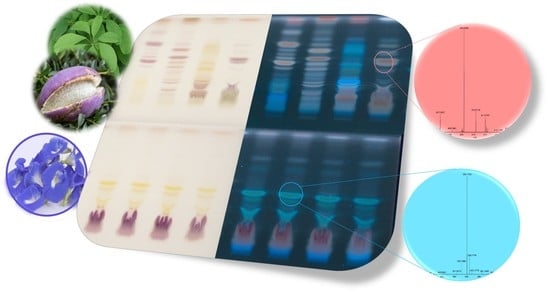
Effect-Directed Profiling of Akebia quinata and Clitoria ternatea via High-Performance Thin-Layer Chromatography, Planar Assays and High-Resolution Mass Spectrometry
Abstract
:1. Introduction
2. Results and Discussion
2.1. Effect-Directed Profiling of Akebia quinata
2.2. Effect-Directed Profiling of Clitoria ternatea
2.3. Agonistic and Antagonistic Endocrine Profiling of A. quinata and C. ternatea
2.4. α-Amylase Effect-Directed Profiling of A. quinata and C. ternatea
3. Materials and Methods
3.1. Chemicals
3.2. Sample Preparation
3.3. Effect-Directed HPTLC Profiling
3.4. HPTLC–Heart-Cut–HPLC–HESI-HRMS
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Maciąg, D.; Dobrowolska, E.; Sharafan, M.; Ekiert, H.; Tomczyk, M.; Szopa, A. Akebia quinata and Akebia trifoliata—A review of phytochemical composition, ethnopharmacological approaches and biological studies. J. Ethnopharmacol. 2021, 280, 114486. [Google Scholar] [CrossRef] [PubMed]
- Kitaoka, F.; Kakiuchi, N.; Long, C.; Itoga, M.; Mitsue, A.; Mouri, C.; Mikage, M. Molecular Characterization of Akebia Plants and the Derived Traditional Herbal Medicine. Biol. Pharm. Bull. 2009, 32, 665–670. [Google Scholar] [CrossRef] [Green Version]
- Ochmian, I.; Guan, T.; Kubus, M. Description and assessment of chemical properties of fruits of the chocolate vine (five-leaf Akebia) Akebia quinata (HOUTT.) Decne and dead man’s fingers Decaisnea insignis (GRIFF.) HOKK.F. & Thomson, grown in Szczecin and in the Arboretum in Glinna (northwestern Poland). J. Elemntology 2014, 19, 1073–1084. [Google Scholar] [CrossRef]
- Rim, A.-R.; Kim, S.-J.; Jeon, K.-I.; Park, E.-J.; Park, H.-R.; Lee, S.-C. Antioxidant Activity of Extracts from Akebia quinata Decne. Prev. Nutr. Food Sci. 2006, 11, 84–87. [Google Scholar] [CrossRef]
- Shin, S.; Son, D.; Kim, M.; Lee, S.; Roh, K.B.; Ryu, D.; Lee, J.; Jung, E.; Park, D. Ameliorating Effect of Akebia quinata Fruit Extracts on Skin Aging Induced by Advanced Glycation End Products. Nutrients 2015, 7, 9337–9352. [Google Scholar] [CrossRef] [Green Version]
- Lee, E.K.; Kwon, W.Y.; Lee, J.W.; Yoon, J.A.; Chung, K.H.; Song, B.C.; An, J.H. Quality Characteristics and Antioxidant Activity of Vinegar Supplemented Added with Akebia quinata fruit during fermentation. J. Korean Soc. Food Sci. Nutr. 2014, 43, 1217–1227. [Google Scholar] [CrossRef]
- Li, X.; Xia, Y.; Li, G.; Zhan, Z.; Yao, R.; Li, M. Traditional uses, phytochemistry, pharmacology, and toxicology of Akebiae Caulis and its synonyms: A review. J. Ethnopharmacol. 2021, 277, 114245. [Google Scholar] [CrossRef]
- Han, S.H.; Kim, Y.W.; Hyun, C. Evaluation of diuretic and hemodynamic effect of extract from Akebia quinata decaisne in dogs. J. Vet. Clin. 2012, 29, 203–206. [Google Scholar]
- Gong, L.-l.; Yang, S.; Liu, H.; Zhang, W.; Ren, L.-l.; Han, F.-f.; Lv, Y.-l.; Wan, Z.-R.; Liu, L.-h. Anti-nociceptive and anti-inflammatory potentials of Akebia saponin D. Eur. J. Pharmacol. 2019, 845, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.; You, Y.; Jun, W. Anti-obesity effects of extracts from young Akebia quinata D. leaves. J. Korean Soc. Food Sci. Nutr. 2014, 43, 200–206. [Google Scholar] [CrossRef]
- Sung, Y.Y.; Kim, D.S.; Kim, H.K. Akebia quinata extract exerts anti-obesity and hypolipidemic effects in high-fat diet-fed mice and 3T3-L1 adipocytes. J. Ethnopharmacol. 2015, 168, 17–24. [Google Scholar] [CrossRef]
- Park, S.H.; Jang, S.; Lee, S.W.; Park, S.D.; Sung, Y.Y.; Kim, H.K. Akebia quinata Decaisne aqueous extract acts as a novel anti-fatigue agent in mice exposed to chronic restraint stress. J. Ethnopharmacol. 2018, 222, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, M.A.; Ko, H.J.; Lee, H.; Aminul Haque, M.; Park, I.S.; Lee, D.S.; Woo, E.R. Oleanane triterpenoids from Akebiae Caulis exhibit inhibitory effects on Aβ42 induced fibrillogenesis. Arch. Pharm. Res. 2017, 40, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.J.; Lee, C.O.; Lee, K.T.; Choi, J.; Park, H.J. Structure-activity relationship of oleanane disaccharides isolated from Akebia quinata versus cytotoxicity against cancer cells and NO inhibition. Biol. Pharm. Bull. 2004, 27, 744–747. [Google Scholar] [CrossRef] [Green Version]
- Suhan, J.; Lee, S.H.; Song, Y.S.; Lee, S.Y.; Kim, S.Y.; Ko, K.S. Effect of beverage containing fermented akebia quinata extracts on alcoholic hangover. Prev. Nutr. Food Sci. 2016, 21, 9–13. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Ren, H.; Xu, Q.L.; Zhou, Z.Y.; Wu, P.; Wei, X.Y.; Cao, Y.; Chen, X.X.; Tan, J.W. Antibacterial oleanane-type triterpenoids from pericarps of Akebia trifoliata. Food Chem. 2015, 168, 623–629. [Google Scholar] [CrossRef]
- Yu, N.; Wang, X.; Ning, F.; Jiang, C.; Li, Y.; Peng, H.; Xiong, H. Development of antibacterial pectin from Akebia trifoliata var. australis waste for accelerated wound healing. Carbohydr. Polym. 2019, 217, 58–68. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Kumar, V.; Kumar, N.S.; Heinrich, M. The Ayurvedic medicine Clitoria ternatea-From traditional use to scientific assessment. J. Ethnopharmacol. 2008, 120, 291–301. [Google Scholar]
- Adisakwattana, S.; Pasukamonset, P.; Chusak, C. Clitoria ternatea beverages and antioxidant usage. In Pathology; Academic Press: Cambridge, MA, USA, 2020; pp. 189–196. [Google Scholar] [CrossRef]
- Al-Snafi, A.E. Pharmacological importance of Clitoria ternatea-A review. IOSR J. Pharm. 2016, 6, 68–83. [Google Scholar]
- Dey, A.; De, J.N. Traditional use of plants against snakebite in indian subcontinent: A review of the recent literature. Afr. J. Tradit. Complement. Altern. Med. 2012, 9, 153–174. [Google Scholar] [CrossRef] [Green Version]
- Bharathee, R.C.; Ranjith, P.; Chandana, A.K.; Chandra Jayakody, J.R.A.; Daya, R.W. In vitro antirheumatoid arthritic activity of aqueous of aqueous root extract of Clitoria ternatea. Int. Res. J. Pharm. 2014, 5, 926–928. [Google Scholar] [CrossRef]
- Swathi, K.P.; Jayaram, S.; Sugumar, D.; Rymbai, E. Evaluation of anti-inflammatory and anti-arthritic property of ethanolic extract of Clitoria ternatea. Chin. Herb. Med. 2021, 13, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Damodaran, T.; Cheah, P.S.; Murugaiyah, V.; Hassan, Z. The nootropic and anticholinesterase activities of Clitoria ternatea Linn. root extract: Potential treatment for cognitive decline. Neurochem. Int. 2020, 139, 104785. [Google Scholar] [CrossRef] [PubMed]
- Rai, K.S.; Murthy, K.D.; Karanth, K.S.; Nalini, K.; Rao, M.S.; Srinivasan, K.K. Clitoria ternatea root extract enhances acetylcholine content in rat hippocampus. Fitoterapia 2002, 73, 685–689. [Google Scholar] [CrossRef]
- Taur, D.J.; Patil, R.Y. Evaluation of antiasthmatic activity of Clitoria ternatea L. roots. J. Ethnopharmacol. 2011, 136, 374–376. [Google Scholar] [CrossRef]
- Talpate, K.A.; Bhosale, U.A.; Zambare, M.R. Clitorea ternatea, a herb from Indian folklore, improves streptozotocin-induced diabetes and diabetes-induced cognitive decline in rats. J. Chinese Integr. Med. 2012, 10, 939–947. [Google Scholar] [CrossRef]
- Talpate, K.; Bhosale, U.; Zambare, M.; Somani, R. Neuroprotective and nootropic activity of Clitorea ternatea Linn.(Fabaceae) leaves on diabetes induced cognitive decline in experimental animals. J. Pharm. Bioallied Sci. 2014, 6, 48–55. [Google Scholar] [CrossRef]
- Shyamkumar, I.B.; Ishwar, B. Anti inflammatory, analgesic and phytochemical studies of Clitoria ternatea linn flower extract. Int. Res. J. Pharm. 2012, 3, 208–210. [Google Scholar]
- Escher, G.B.; Marques, M.B.; do Carmo, M.A.V.; Azevedo, L.; Furtado, M.M.; Sant’Ana, A.S.; da Silva, M.C.; Genovese, M.I.; Wen, M.; Zhang, L.; et al. Clitoria ternatea L. petal bioactive compounds display antioxidant, antihemolytic and antihypertensive effects, inhibit α-amylase and α-glucosidase activities and reduce human LDL cholesterol and DNA induced oxidation. Food Res. Int. 2020, 128, 108763. [Google Scholar] [CrossRef]
- Bhosale, U.; Zambare, M.; Somani, R.; Talpate, K. Antihyperglycemic and antioxidant activity of Clitorea ternatea Linn. on streptozotocin-induced diabetic rats. AYU (An Int. Q. J. Res. Ayurveda) 2013, 34, 433–439. [Google Scholar] [CrossRef] [Green Version]
- Indrianingsih, A.W.; Wulanjati, M.P.; Windarsih, A.; Bhattacharjya, D.K.; Suzuki, T.; Katayama, T. In vitro studies of antioxidant, antidiabetic, and antibacterial activities of Theobroma cacao, Anonna muricata and Clitoria ternatea. Biocatal. Agric. Biotechnol. 2021, 33, 101995. [Google Scholar] [CrossRef]
- Kamilla, L.; Mnsor, S.M.; Ramanathan, S.; Sasidharan, S. Antimicrobial activity of Clitoria ternatea (L.) extracts. Pharmacologyonline 2009, 1, 731–738. [Google Scholar]
- Mahmad, N.; Taha, R.M.; Othman, R.; Abdullah, S.; Anuar, N.; Elias, H.; Rawi, N. Anthocyanin as potential source for antimicrobial activity in Clitoria ternatea L. and Dioscorea alata L. Pigment Resin Technol. 2018, 47, 490–495. [Google Scholar] [CrossRef]
- Niranjan, M.; Vaishnav, V.; Mankar, P. In-vitro analysis of antioxidant and antimicrobial properties of Garcinia mangostana L. (pericarp) and Clitoria ternatea (flower). Pharma Innov. J. 2020, 9, 468–472. [Google Scholar]
- Dhanasekaran, S.; Rajesh, A.; Mathimani, T.; Melvin Samuel, S.; Shanmuganathan, R.; Brindhadevi, K. Efficacy of crude extracts of Clitoria ternatea for antibacterial activity against gram negative bacterium (Proteus mirabilis). Biocatal. Agric. Biotechnol. 2019, 21, 101328. [Google Scholar] [CrossRef]
- Prasansuklab, A.; Theerasri, A.; Rangsinth, P.; Sillapachaiyaporn, C.; Chuchawankul, S.; Tencomnao, T. Anti-COVID-19 drug candidates: A review on potential biological activities of natural products in the management of new coronavirus infection. J. Tradit. Complement. Med. 2021, 11, 144–157. [Google Scholar]
- Nair, V.; Bang, W.Y.; Schreckinger, E.; Andarwulan, N.; Cisneros-Zevallos, L. Protective Role of Ternatin Anthocyanins and Quercetin Glycosides from Butterfly Pea (Clitoria ternatea Leguminosae) Blue Flower Petals against Lipopolysaccharide (LPS)-Induced Inflammation in Macrophage Cells. J. Agric. Food Chem. 2015, 63, 6355–6365. [Google Scholar] [CrossRef]
- Makasana, J.; Dholakiya, B.Z.; Gajbhiye, N.A.; Raju, S. Extractive determination of bioactive flavonoids from butterfly pea (Clitoria ternatea Linn.). Res. Chem. Intermed. 2017, 43, 783–799. [Google Scholar] [CrossRef]
- Lee, D.; Lee, J.S.; Sezirahiga, J.; Kwon, H.C.; Jang, D.S.; Kang, K.S. Bioactive phytochemicals isolated from akebia quinata enhances glucose-stimulated insulin secretion by inducing pdx-1. Plants 2020, 9, 1087. [Google Scholar] [CrossRef]
- Terahara, N.; Oda, M.; Matsui, T.; Osajima, Y.; Saito, N.; Toki, K.; Honda, T. Five new anthocyanins, ternatins A3, B4, B3, B2, and D2, from Clitoria ternatea flowers. J. Nat. Prod. 1996, 59, 139–144. [Google Scholar] [CrossRef]
- Rajamanickam, M. Isolation and Characterizations of new alkaloid 3-deoxy- 3, 11-epoxy cephalotaxine from Clitoria ternatea. J. Drug Deliv. Ther. 2019, 9, 458–462. [Google Scholar] [CrossRef]
- Mimaki, Y.; Doi, S.; Kuroda, M.; Yokosuka, A. Triterpene glycosides from the stems of Akebia quinata. Chem. Pharm. Bull. 2007, 55, 1319–1324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ko, H.J.; Lee, J.H.; Kim, Y.S.; Lee, J.H.; Woo, E.R. A new triterpene glycoside from the stems of Akebia quinata. Bull. Korean Chem. Soc. 2015, 36, 356–359. [Google Scholar] [CrossRef]
- Wu, F.S.; Hung, C.J.; Lin, C.L.; Chang, T.H.; Chen, C.L.; Sung, P.J.; Cheng, M.J.; Huang, H.Y.; Chen, J.J. New Norneolignan and Bioactive Constituents of Clitoria ternatea. Chem. Nat. Compd. 2020, 56, 1000–1004. [Google Scholar] [CrossRef]
- Escher, G.B.; Wen, M.; Zhang, L.; Rosso, N.D.; Granato, D. Phenolic composition by UHPLC-Q-TOF-MS/MS and stability of anthocyanins from Clitoria ternatea L. (butterfly pea) blue petals. Food Chem. 2020, 331, 127341. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Yang, S.; Li, J.; Ouyang, H.; He, M.; Feng, Y.; Tan, T. A four-step filtering strategy based on ultra-high-performance liquid chromatography coupled to quadrupole-time-of-flight tandem mass spectrometry for comprehensive profiling the major chemical constituents of Akebiae Fructus. Rapid Commun. Mass Spectrom. 2019, 33, 1464–1474. [Google Scholar] [CrossRef] [PubMed]
- Ling, Y.; Zhang, Y.; Zhou, Y.; Jiang, D.; Xu, L.; Liao, L. Rapid detection and characterization of the major chemical constituents in Akebia quinata by high performance liquid chromatography coupled to electrospray ionization and quadrupole time-of-flight mass spectrometry. Anal. Methods 2016, 8, 2634–2644. [Google Scholar] [CrossRef]
- Jin, H.G.; Kim, A.R.; Ko, H.J.; Lee, S.K.; Woo, E.R. Three new lignan glycosides with IL-6 inhibitory activity from Akebia quinata. Chem. Pharm. Bull. 2014, 62, 288–293. [Google Scholar] [CrossRef] [Green Version]
- Mahadeva Rao, U.S.; Zin, T.; Abdurrazakb, M.; Ahmad, A.A. Chemistry and pharmacology of syringin, a novel bioglycoside: A review. Asian J. Pharm. Clin. Res. 2015, 8, 20–25. [Google Scholar]
- Sun, J.; Feng, Y.; Wang, Y.; Ji, Q.; Cai, G.; Shi, L.; Wang, Y.; Huang, Y.; Zhang, J.; Li, Q. α-hederin induces autophagic cell death in colorectal cancer cells through reactive oxygen species dependent AMPK/mTOR signaling pathway activation. Int. J. Oncol. 2019, 54, 1601–1612. [Google Scholar] [CrossRef] [Green Version]
- Ayeleso, T.B.; Matumba, M.G.; Mukwevho, E. Oleanolic acid and its derivatives: Biological activities and therapeutic potential in chronic diseases. Molecules 2017, 22, 1915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alakurtti, S.; Mäkelä, T.; Koskimies, S.; Yli-Kauhaluoma, J. Pharmacological properties of the ubiquitous natural product betulin. Eur. J. Pharm. Sci. 2006, 29, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xu, Q.L.; Zheng, M.F.; Ren, H.; Lei, T.; Wu, P.; Zhou, Z.Y.; Wei, X.Y.; Tan, J.W. Bioactive 30-noroleanane triterpenes from the pericarps of akebia trifoliata. Molecules 2014, 19, 4301–4312. [Google Scholar] [CrossRef] [Green Version]
- Yu, S.; Liu, M.; Gu, X.; Ding, F. Neuroprotective effects of salidroside in the PC12 cell model exposed to hypoglycemia and serum limitation. Cell. Mol. Neurobiol. 2008, 28, 1067–1078. [Google Scholar] [CrossRef] [PubMed]
- Mus, A.A.; Goh, L.P.W.; Marbawi, H.; Gansau, J.A. The Biosynthesis and Medicinal Properties of Taraxerol. Biomedicines 2022, 10, 807. [Google Scholar] [CrossRef]
- Gong, L.; Feng, D.; Wang, T.; Ren, Y.; Liu, Y.; Wang, J. Inhibitors of α-amylase and α-glucosidase: Potential linkage for whole cereal foods on prevention of hyperglycemia. Food Sci. Nutr. 2020, 8, 6320–6337. [Google Scholar] [CrossRef]
- Nikolaichuk, H.; Choma, I.M.; Morlock, G.E. Bioactivity Profiles on 15 Different Effect Mechanisms for 15 Golden Root Products via High-Performance Thin-Layer Chromatography, Planar Assays, and High-Resolution Mass Spectrometry. Molecules 2023, 28, 1535. [Google Scholar] [CrossRef]
- Klingelhöfer, I.; Hockamp, N.; Morlock, G.E. Non-targeted detection and differentiation of agonists versus antagonists, directly in bioprofiles of everyday products. Anal. Chim. Acta 2020, 1125, 288–298. [Google Scholar] [CrossRef]
- Morlock, G.E.; Heil, J.; Bardot, V.; Lenoir, L.; Cotte, C.; Dubourdeaux, M. Effect-directed profiling of 17 different fortified plant extracts by high-performance thin-layer chromatography combined with six planar assays and high-resolution mass spectrometry. Molecules 2021, 26, 1468. [Google Scholar] [CrossRef]
- Schreiner, T.; Sauter, D.; Friz, M.; Heil, J.; Morlock, G.E. Is Our Natural Food Our Homeostasis? Array of a Thousand Effect-Directed Profiles of 68 Herbs and Spices. Front. Pharmacol. 2021, 12, 755941. [Google Scholar] [CrossRef]
- Jamshidi-Aidji, M.; Morlock, G.E. From bioprofiling and characterization to bioquantification of natural antibiotics by direct bioautography linked to high-resolution mass spectrometry: Exemplarily shown for Salvia miltiorrhiza root. Anal. Chem. 2016, 88, 10979–10986. [Google Scholar] [CrossRef] [PubMed]
- Meyer, D.; Marin-Kuan, M.; Debon, E.; Serrant, P.; Cottet-Fontannaz, C.; Schilter, B.; Morlock, G.E. Detection of low levels of genotoxic compounds in food contact materials using an alternative HPTLC-SOS-Umu-C assay. ALTEX 2021, 38, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Debon, E.; Rogeboz, P.; Latado, H.; Morlock, G.E.; Meyer, D.; Cottet-Fontannaz, C.; Scholz, G.; Schilter, B.; Marin-Kuan, M. Incorporation of Metabolic Activation in the HPTLC-SOS-Umu-C Bioassay to Detect Low Levels of Genotoxic Chemicals in Food Contact Materials. Toxics 2022, 10, 501. [Google Scholar] [CrossRef] [PubMed]
- Meyer, D.; Marin-Kuan, M.; Mayrhofer, E.; Kirchnawy, C.; Debon, E.; Latado, H.; Patin, A.; Schilter, B.; Morlock, G.E. Effect-detection by planar SOS-Umu-C genotoxicity bioassay and chemical identification of genotoxins in packaging migrates, proven by microtiter plate assays SOS-Umu-C and Ames-MPF. Food Control 2023, 147, 109546. [Google Scholar] [CrossRef]
- Azadniya, E.; Morlock, G.E. Automated piezoelectric spraying of biological and enzymatic assays for effect-directed analysis of planar chromatograms. J. Chromatogr. A 2019, 1602, 458–466. [Google Scholar] [CrossRef] [PubMed]
- Jamshidi-Aidji, M.; Morlock, G.E. Fast Equivalency Estimation of Unknown Enzyme Inhibitors in Situ the Effect-Directed Fingerprint, Shown for Bacillus Lipopeptide Extracts. Anal. Chem. 2018, 90, 14260–14268. [Google Scholar] [CrossRef]
- Mehl, A.; Schwack, W.; Morlock, G.E. On-surface autosampling for liquid chromatography−mass spectrometry. J. Chromatogr. A 2021, 1651, 462334. [Google Scholar] [CrossRef]




| Zone | hRF | Formula | Calculated Mass [Da] | Observed Mass [Da] | Adduct Ions | Mass Error (ppm) | Tentative Assignment |
| I | 52 | C17H24O9 | 372.1420 | 431.1563 395.1308 | [M+CH3COO]− [M+Na]+ | −1.03 1.07 | Syringin |
| C14H20O8 | 316.1158 | 315.1082 339.1054 | [M−H]− [M+Na]+ | 1.03 −1.12 | Vanilloloside | ||
| C14H20O7 | 300.1209 | 299.1132 323.1097 | [M−H]− [M+Na]+ | 1.42 1.30 | Salidroside | ||
| II | 60 | C41H66O12 | 750.4554 | 749.4489 773.4433 | [M−H]− [M+Na]+ | −1.04 1.71 | α-Hederin |
| C24H40O11 | 504.2571 | 527.2451 | [M+Na]+ | 2.32 | Cuneataside E | ||
| III | 65 | C30H50O2 | 442.3811 | 465.3706 | [M+Na]+ | −0.60 | Betulin |
| C30H48O3 | 456.3604 | 479.3498 | [M+Na]+ | −0.37 | Oleanolic acid | ||
| IV | 55 | C14H20O7 | 300.1209 | 299.1134 323.1095 | [M−H]− [M+Na]+ | 0.63 1.92 | Salidroside |
| C29H44O4 | 456.3240 | 455.3170 479.3138 | [M−H]− [M+Na]+ | −0.61 −1.21 | Quinatic acid |
| Zone | hRF | Formula | Calculated Mass [Da] | Observed Mass [Da] | Adduct Ions | Mass Error (ppm) | Tentative Assignment |
| I | 46 | C30H50O | 426.3862 | 449.3751 | [M+Na]+ | 0.69 | Taraxerol |
| C27H30O15 | 594.1585 | 593.1525 617.1483 | [M−H]− [M+Na]+ | −2.13 −0.91 | Kaempferol-3-rutinoside | ||
| C27H30O16 | 610.1534 | 609.1473 633.1435 | [M−H]− [M+Na]+ | −1.91 −1.34 | Quercetin-3-rutinoside | ||
| II | 56 | C18H34O2 | 282.2569 | 281.2489 305.2453 | [M−H]− [M+Na]+ | −0.95 −0.59 | Octadecenoic acid |
| C21H20O11 | 448.1006 | 447.0937 471.0902 | [M−H]− [M+Na]+ | −0.89 −0.85 | Kaempferol-3-glucoside |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nikolaichuk, H.; Choma, I.M.; Morlock, G.E. Effect-Directed Profiling of Akebia quinata and Clitoria ternatea via High-Performance Thin-Layer Chromatography, Planar Assays and High-Resolution Mass Spectrometry. Molecules 2023, 28, 2893. https://doi.org/10.3390/molecules28072893
Nikolaichuk H, Choma IM, Morlock GE. Effect-Directed Profiling of Akebia quinata and Clitoria ternatea via High-Performance Thin-Layer Chromatography, Planar Assays and High-Resolution Mass Spectrometry. Molecules. 2023; 28(7):2893. https://doi.org/10.3390/molecules28072893
Chicago/Turabian StyleNikolaichuk, Hanna, Irena M. Choma, and Gertrud E. Morlock. 2023. "Effect-Directed Profiling of Akebia quinata and Clitoria ternatea via High-Performance Thin-Layer Chromatography, Planar Assays and High-Resolution Mass Spectrometry" Molecules 28, no. 7: 2893. https://doi.org/10.3390/molecules28072893
APA StyleNikolaichuk, H., Choma, I. M., & Morlock, G. E. (2023). Effect-Directed Profiling of Akebia quinata and Clitoria ternatea via High-Performance Thin-Layer Chromatography, Planar Assays and High-Resolution Mass Spectrometry. Molecules, 28(7), 2893. https://doi.org/10.3390/molecules28072893