Lower Salinomycin Concentration Increases Apoptotic Detachment in High-Density Cancer Cells
Abstract
:1. Introduction
2. Results and Discussion
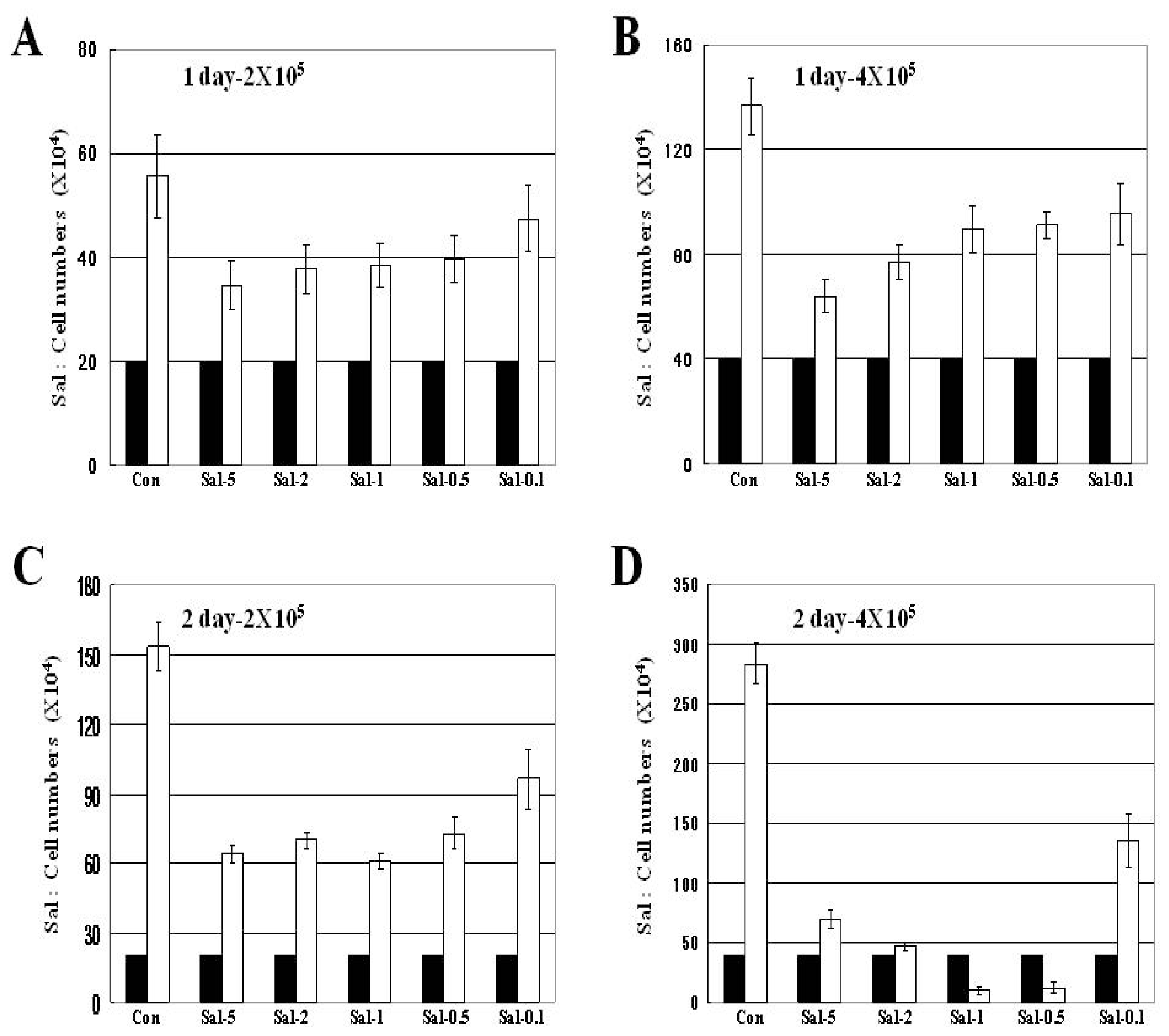
2.1. Attached Cells in High Density Culture are more Effectively Reduced by Longer Sal Exposure
2.2. Treatment with 0.5 μM Sal Reduces the Number of Attached Cells Better than 5 μM Sal in High Density Cultures with Longer Exposure
2.3. Treatment with 0.5 μM Sal Increases Easily-Detachable Cells in High Density Culture
2.4. Treatment with 0.5 μM Sal Increases Proliferation in High Density Culture, but not Differentiation for Attachment
2.5. Treatment with 0.5 μM Sal Markedly Increases Apoptotic Cells in High Density Culture
2.6. Treatment with 0.5 μM Sal may Increase Cellular Toxicity in High Density Cultures
2.7. Space among Cells Contributes to Increased Detachment in 0.5 μM Sal Treatment
3. Discussion
4. Experimental Section
4.1. Reagents
4.2. Cell Culturing
4.3. Cell Counting
4.4. Live Cell Imaging
4.5. Cell Viability Assay
4.6. Fluorescence-Activated Cell Sorting (FACS) Analysis
4.7. Annexin V Analysis
4.8. Terminal Transferase dUTP Nick End Labeling (TUNEL) Assay
4.9. Cellular Rho Level Tests
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
ijms-13-13169-s001.pdfAcknowledgments
References
- Miyazaki, Y.; Shibuya, M.; Sugawara, H.; Kawaguchi, O.; Hirsoe, C. Salinomycin, a new polyether antibiotic. J. Antibio (Tokyo) 1974, 27, 814–821. [Google Scholar]
- Mitani, M.; Yamanishi, T.; Miyazaki, Y. Salinomycin: A new monovalent cation ionophore. Biochem. Biophys. Res. Commun 1975, 66, 1231–1236. [Google Scholar]
- Mitani, M.; Yamanishi, T.; Miyazaki, Y.; Otake, N. Salinomycin effects on mitochondrial ion translocation and respiration. Antimicrob. Agents Chemother 1976, 9, 655–660. [Google Scholar]
- Mahmoudi, N.; de Julian-Ortiz, J.V.; Ciceron, L.; Galvez, J.; Mazier, D.; Danis, M.; Derouin, F.; Garcia-Domenech, R. Identification of new antimalarial drugs by linear discriminant analysis and topological virtual screening. J. Antimicrob. Chemother 2006, 57, 489–497. [Google Scholar]
- Bardsley, M.R.; Horvath, V.J.; Asuzu, D.T.; Lorincz, A.; Redelman, D.; Hayashi, Y.; Popko, L.N.; Young, D.L.; Lomberk, G.A.; Urrutia, R.A.; et al. Kitlow stem cells cause resistance to Kit/platelet-derived growth factor alpha inhibitors in murine gastrointestinal stromal tumors. Gastroenterology 2010, 139, 942–952. [Google Scholar]
- Dong, T.T.; Zhou, H.M.; Wang, L.L.; Feng, B.; Lv, B.; Zheng, M.H. Salinomycin selectively targets “CD133+” cell subpopulations and decreases malignant traits in colorectal cancer lines. Ann. Surg. Oncol 2011, 18, 1797–1804. [Google Scholar]
- Fuchs, D.; Heinold, A.; Opelz, G.; Daniel, V.; Naujokat, C. Salinomycin induces apoptosis and overcomes apoptosis resistance in human cancer cells. Biochem. Biophys. Res. Commun 2009, 390, 743–749. [Google Scholar]
- Gong, C.; Yao, H.; Liu, Q.; Chen, J.; Shi, J.; Su, F.; Song, E. Markers of tumor-initiating cells predict chemoresistance in breast cancer. PLoS One 2010, 5, e15630. [Google Scholar]
- Gupta, P.B.; Onder, T.T.; Jiang, G.; Tao, K.; Kuperwasser, C.; Weinberg, R.A.; Lander, E.S. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 2009, 138, 645–659. [Google Scholar]
- Huczynski, A. Salinomycin: A new cancer drug candidate. Chem. Biol. Drug Des 2012, 79, 235–238. [Google Scholar]
- Ketola, K.; Hilvo, M.; Hyotylainen, T.; Vuoristo, A.; Ruskeepaa, A.L.; Oresic, M.; Kallioniemi, O.; Iljin, K. Salinomycin inhibits prostate cancer growth and migration via induction of oxidative stress. Br. J. Cancer 2012, 106, 99–106. [Google Scholar]
- Kim, K.Y.; Yu, S.N.; Lee, S.Y.; Chun, S.S.; Choi, Y.L.; Park, Y.M.; Song, C.S.; Chatterjee, B.; Ahn, S.C. Salinomycin-induced apoptosis of human prostate cancer cells due to accumulated reactive oxygen species and mitochondrial membrane depolarization. Biochem. Biophys. Res. Commun 2011, 413, 80–86. [Google Scholar]
- Lu, D.; Choi, M.Y.; Yu, J.; Castro, J.E.; Kipps, T.J.; Carson, D.A. Salinomycin inhibits Wnt signaling and selectively induces apoptosis in chronic lymphocytic leukemia cells. Proc. Natl. Acad. Sci. USA 2011, 108, 13253–13257. [Google Scholar]
- Tang, Q.L.; Zhao, Z.Q.; Li, J.C.; Liang, Y.; Yin, J.Q.; Zou, C.Y.; Xie, X.B.; Zeng, Y.X.; Shen, J.N.; Kang, T.; et al. Salinomycin inhibits osteosarcoma by targeting its tumor stem cells. Cancer Lett 2011, 311, 113–121. [Google Scholar]
- Wang, Y. Effects of salinomycin on cancer stem cell in human lung adenocarcinoma A549 cells. Med. Chem 2011, 7, 106–111. [Google Scholar]
- Zhang, G.N.; Liang, Y.; Zhou, L.J.; Chen, S.P.; Chen, G.; Zhang, T.P.; Kang, T.; Zhao, Y.P. Combination of salinomycin and gemcitabine eliminates pancreatic cancer cells. Cancer Lett 2011, 313, 137–144. [Google Scholar]
- Zhi, Q.M.; Chen, X.H.; Ji, J.; Zhang, J.N.; Li, J.F.; Cai, Q.; Liu, B.Y.; Gu, Q.L.; Zhu, Z.G.; Yu, Y.Y. Salinomycin can effectively kill ALDH (high) stem-like cells on gastric cancer. Biomed. Pharmacother 2011, 65, 509–515. [Google Scholar]
- Fuchs, D.; Daniel, V.; Sadeghi, M.; Opelz, G.; Naujokat, C. Salinomycin overcomes ABC transporter-mediated multidrug and apoptosis resistance in human leukemia stem cell-like KG-1a cells. Biochem. Biophys. Res. Commun 2010, 394, 1098–1104. [Google Scholar]
- Kim, W.K.; Kim, J.H.; Yoon, K.; Kim, S.; Ro, J.; Kang, H.S.; Yoon, S. Salinomycin, a p-glycoprotein inhibitor, sensitizes radiation-treated cancer cells by increasing DNA damage and inducing G2 arrest. Invest. New Drugs 2012, 30, 1311–1318. [Google Scholar]
- Riccioni, R.; Dupuis, M.L.; Bernabei, M.; Petrucci, E.; Pasquini, L.; Mariani, G.; Cianfriglia, M.; Testa, U. The cancer stem cell selective inhibitor salinomycin is a p-glycoprotein inhibitor. Blood Cells Mol. Dis 2010, 45, 86–92. [Google Scholar]
- Kim, J.H.; Chae, M.; Kim, W.K.; Kim, Y.J.; Kang, H.S.; Kim, H.S.; Yoon, S. Salinomycin sensitizes cancer cells to the effects of doxorubicin and etoposide treatment by increasing DNA damage and reducing p21 protein. Br. J. Pharmacol 2011, 162, 773–784. [Google Scholar]
- Kim, J.H.; Yoo, H.I.; Kang, H.S.; Ro, J.; Yoon, S. Salinomycin sensitizes antimitotic drugs-treated cancer cells by increasing apoptosis via the prevention of G2 arrest. Biochem. Biophys. Res. Commun 2012, 418, 98–103. [Google Scholar]
- Fang, Y.; Sullivan, R.; Graham, C.H. Confluence-dependent resistance to doxorubicin in human MDA-MB-231 breast carcinoma cells requires hypoxia-inducible factor-1 activity. Exp. Cell Res 2007, 313, 867–877. [Google Scholar]
- Menard, R.E.; Jovanovski, A.P.; Mattingly, R.R. Active p21-activated kinase 1 rescues MCF10A breast epithelial cells from undergoing anoikis. Neoplasia 2005, 7, 638–645. [Google Scholar]
- Lagas, J.S.; Sparidans, R.W.; van Waterschoot, R.A.; Wagenaar, E.; Beijnen, J.H.; Schinkel, A.H. P-glycoprotein limits oral availability, brain penetration, and toxicity of an anionic drug, the antibiotic salinomycin. Antimicrob. Agents Chemother 2008, 52, 1034–1039. [Google Scholar]
- Johnson, L.V.; Walsh, M.L.; Chen, L.B. Localization of mitochondria in living cells with rhodamine 123. Proc. Natl. Acad. Sci. USA 1980, 77, 990–994. [Google Scholar]
- Lachowiez, R.M.; Clayton, B.; Thallman, K.; Dix, J.A.; van Buskirk, R.G. Rhodamine 123 as a probe of in vitro toxicity in MDCK cells. Cytotechnology 1989, 2, 203–211. [Google Scholar]
- Boehmerle, W.; Endres, M. Salinomycin induces calpain and cytochrome c-mediated neuronal cell death. Cell Death Dis 2011, 2, e168. [Google Scholar]
- Kim, J.H.; Kim, T.H.; Kang, H.S.; Ro, J.; Kim, H.S.; Yoon, S. SP600125, an inhibitor of Jnk pathway, reduces viability of relatively resistant cancer cells to doxorubicin. Biochem. Biophys. Res. Commun 2009, 387, 450–455. [Google Scholar]
- Kim, J.H.; Lee, S.C.; Ro, J.; Kang, H.S.; Kim, H.S.; Yoon, S. Jnk signaling pathway-mediated regulation of Stat3 activation is linked to the development of doxorubicin resistance in cancer cell lines. Biochem. Pharmacol 2010, 79, 373–380. [Google Scholar]
- Ha, G.H.; Baek, K.H.; Kim, H.S.; Jeong, S.J.; Kim, C.M.; McKeon, F.; Lee, C.W. p53 activation in response to mitotic spindle damage requires signaling via BubR1-mediated phosphorylation. Cancer Res 2007, 67, 7155–7164. [Google Scholar]



© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kim, J.-H.; Kim, T.Y.; Kim, H.S.; Hong, S.; Yoon, S. Lower Salinomycin Concentration Increases Apoptotic Detachment in High-Density Cancer Cells. Int. J. Mol. Sci. 2012, 13, 13169-13182. https://doi.org/10.3390/ijms131013169
Kim J-H, Kim TY, Kim HS, Hong S, Yoon S. Lower Salinomycin Concentration Increases Apoptotic Detachment in High-Density Cancer Cells. International Journal of Molecular Sciences. 2012; 13(10):13169-13182. https://doi.org/10.3390/ijms131013169
Chicago/Turabian StyleKim, Ju-Hwa, Tae Young Kim, Hyung Sik Kim, Suntaek Hong, and Sungpil Yoon. 2012. "Lower Salinomycin Concentration Increases Apoptotic Detachment in High-Density Cancer Cells" International Journal of Molecular Sciences 13, no. 10: 13169-13182. https://doi.org/10.3390/ijms131013169



