Identification and Characterization of 40 Isolated Rehmannia glutinosa MYB Family Genes and Their Expression Profiles in Response to Shading and Continuous Cropping
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification of the MYB Gene Family in R. glutinosa
2.2. Protein Motif and Structure Analysis of the MYBs in R. glutinosa
| Gene Name | Gene ID | Gene Length/bp | Amino Acid Length | Blast Results (Query Cover, e-Value, Identities, Accession No., Description, (Species)) |
|---|---|---|---|---|
| RgMYB1 | CL429Contig1 | 1126 | 273 | 95%, 2 × 10−144, 76%, XP_011070362.1, transcription repressor MYB5-like (Sesamum indicum) |
| RgMYB2 | CL8750Contig1 | 831 | 210 | 90%, 2 × 10−95, 73%, AGN52078.1, MYB-related transcription factor (Salvia miltiorrhiza) |
| RgMYB3 | Unigene7708 | 1036 | 318 | 100%, 0.0, 79%, AGN52106.1, MYB-related transcription factor (S. miltiorrhiza) |
| RgMYB4 | CL128Contig2 | 1582 | 418 | 100%, 0.0, 70%, XP_011087570.1, transcription factor MYB86-like (S. indicum) |
| RgMYB5 | Unigene19474 | 673 | 199 | 95%, 2 × 10−89, 72%, XP_011079754.1, transcription factor MYB114-like (S. indicum) |
| RgMYB6 | Unigene18939 | 897 | 267 | 97%, 1 × 10−96, 59%, AGZ16400.1 MYB7 (Scutellaria baicalensis) |
| RgMYB7 | CL7757Contig1 | 1207 | 306 | 100%, 8 × 10−118, 61%, XP_011086359.1, MYB-related protein Zm1 (S. indicum) |
| RgMYB8 | Unigene14277 | 1100 | 299 | 100%, 4 × 10−161, 73%, XP_011098622.1, MYB-related protein 306-like (S. indicum) |
| RgMYB9 | Unigene7159 | 1338 | 319 | 100%, 1 × 10−153, 74%, XP_011071639.1, MYB-related protein 306-like (S. indicum) |
| RgMYB10 | Unigene16684 | 1017 | 247 | 100%, 4 × 10−92, 63%, XP_011080041.1, MYB-related protein Myb4-like (S. indicum) |
| RgMYB11 | Unigene9736 | 1347 | 251 | 100%, 4 × 10−149, 83%, XP_011091315.1, MYB-related protein Myb4-like (S. indicum) |
| RgMYB12 | Unigene12270 | 862 | 265 | 99%, 3 × 10−71, 52%, XP_011091328.1, MYB-related protein Myb4-like (S. indicum) |
| RgMYB13 | Unigene12354 | 1307 | 292 | 97%, 5 × 10−95, 56%, XP_011100326.1, MYB-related protein Myb4-like (S. indicum) |
| RgMYB14 | Unigene21429 | 1227 | 293 | 98%, 1 × 10−105, 59%, XP_011100326.1, MYB-related protein Myb4-like (S. indicum) |
| RgMYB15 | CL5174Contig3 | 1101 | 219 | 94%, 2 × 10−84, 68%, XP_011089822.1, transcription factor MYB48-like (S. indicum) |
| RgMYB16 | CL9333Contig1 | 1327 | 286 | 100%, 4 × 10−139, 75%, XP_011076599.1, transcription factor MYB108-like (S. indicum) |
| RgMYB17 | Unigene5899 | 1217 | 288 | 96%, 1 × 10−131, 74%, XP_011070788.1, transcription factor MYB24 (S. indicum) |
| RgMYB18 | CL6949Contig1 | 790 | 202 | 91%, 8 × 10−94, 74%, AGN52041.1, MYB-related transcription factor (S. miltiorrhiza) |
| RgMYB26 | CL3700Contig2 | 3518 | 1015 | 100%, 0.0, 75%, XP_011081822.1, MYB-related protein 3R-1 (S. indicum) |
| RgMYB19 | Unigene19140 | 1451 | 419 | 100%, 3 × 10−155, 55%, XP_011076754.1, uncharacterized protein (S. indicum) |
| RgMYB20 | Unigene9854 | 1257 | 303 | 100%, 2 × 10−116, 62%, XP_011088207.1, transcription factor MYB44-like (S. indicum) |
| RgMYB21 | CL9364Contig2 | 1570 | 338 | 100%, 4 × 10−162, 74%, XP_011074411.1, transcription factor MYB44-like (S. indicum) |
| RgMYB22 | CL6089Contig2 | 1303 | 307 | 86%, 1 × 10−142, 70%, XP_011099313.1, transcription factor MYB44-like (S. indicum) |
| RgMYB23 | Unigene11755 | 1536 | 374 | 84%, 6 × 10−161, 80%, XP_011099313.1, transcription factor MYB44-like (S. indicum) |
| RgMYB24 | CL173Contig2 | 1384 | 335 | 100%, 0.0, 79%, XP_011077649.1, transcription factor AS1 (S. indicum) |
| RgMYB25 | Unigene5357 | 1478 | 387 | 96%, 0.0, 84%, XP_011097206.1, protein rough sheath 2 homolog (S. indicum) |
| RgMYB27 | CL5870Contig1 | 2525 | 697 | 99%, 0.0, 74%, XP_011085026.1, telomere repeat-binding protein 4 (S. indicum) |
| RgMYB28 | CL7811Contig2 | 1869 | 593 | 100%, 0.0, 73%, XP_011094643.1, uncharacterized protein (S. indicum) |
| RgMYB29 | Unigene9541 | 836 | 233 | 100%, 3 × 10−105, 69%, XP_011094644.1, uncharacterized protein (S. indicum) |
| RgMYB30 | Unigene18531 | 2173 | 616 | 100%, 0.0, 73%, XP_011079786.1, uncharacterized protein (S. indicum) |
| RgMYB31 | CL4303Contig4 | 1484 | 302 | 100%, 8 × 10−161, 90%, XP_011088080.1, telomere repeat-binding factor 1 (S. indicum) |
| RgMYB32 | Unigene21243 | 1440 | 300 | 100%, 2 × 10−145, 84%, XP_011088080.1, telomere repeat-binding factor 1 (S. indicum) |
| RgMYB33 | CL4502Contig2 | 1359 | 295 | 100%, 2 × 10−126, 75%, XP_011099389.1, telomere repeat-binding factor 1 (S. indicum) |
| RgMYB34 | CL4502Contig3 | 1226 | 294 | 100%, 8 × 10−138, 82%, XP_011099389.1 telomere repeat-binding factor 1 (S. indicum) |
| RgMYB35 | CL3772Contig3 | 1175 | 328 | 100%, 3 × 10−176, 73%, XP_011095766.1, MYB family transcription factor APL (S. indicum) |
| RgMYB36 | Unigene13894 | 1053 | 272 | 68%, 3 × 10−111, 91%, XP_011092595.1, MYB family transcription factor APL (S. indicum) |
| RgMYB37 | CL3105Contig2 | 714 | 187 | 100%, 5 × 10−103, 82%, XP_011097737.1, transcription factor Divaricata-like (S. indicum) |
| RgMYB38 | CL725Contig2 | 1561 | 288 | 100%, 5 × 10−177, 88%, XP_011094729.1, protein REVEILLE 8 (S. indicum) |
| RgMYB39 | CL5749Contig2 | 1086 | 266 | 100%, 6 × 10−133, 83%, XP_011071772.1, transcription factor MYB1R1 (S. indicum) |
| RgMYB40 | CL613Contig1 | 1138 | 260 | 100%, 2 × 10−111, 72%, XP_011071772.1, transcription factor MYB1R1 (S. indicum) |


2.3. Phylogenetic Analysis of the MYB Proteins

2.4. Differential Expression of R. glutinosa MYB Genes in Various Tissues/Developmental Stages

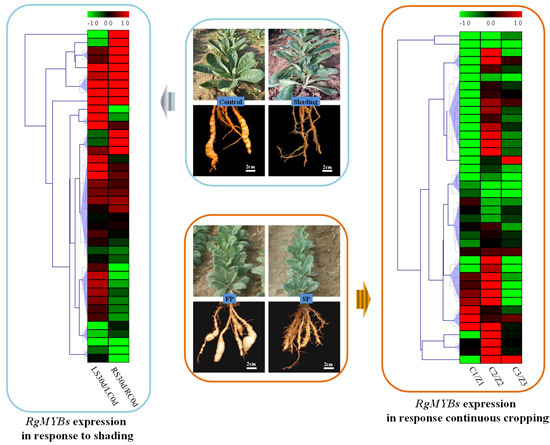
2.5. Expression Profiling of MYB Genes under Shading and Continuous Cropping Stresses

2.6. Discussion
3. Experimental Section
3.1. Plant Material and Sample Collection
3.2. Identification of Putative MYB mRNAs
3.3. Protein Structure and Phylogenetic Analysis
3.4. Expression Profile Analysis of MYB Genes
3.5. Quantitative Real-Time Polymerase Chain Reaction (PCR) Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Klempnauer, K.-H.; Gonda, T.J.; Bishop, J.M. Nucleotide sequence of the retroviral leukemia gene v-MYB and its cellular progenitor c-MYB: The architecture of a transduced oncogene. Cell 1982, 31, 453–463. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Rabinowicz, P.D.; Braun, E.L.; Wolfe, A.D.; Bowen, B.; Grotewold, E. Maize R2R3 Myb genes: Sequence analysis reveals amplification in the higher plants. Genetics 1999, 153, 427–444. [Google Scholar]
- Paz-Ares, J.; Ghosal, D.; Wienand, U.; Peterson, P.A.; Saedler, H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 1987, 6, 3553–3558. [Google Scholar] [PubMed]
- Jin, H.; Martin, C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 1999, 41, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Rosinski, J.A.; Atchley, W.R. Molecular evolution of the MYB family of transcription factors: Evidence for polyphyletic origin. J. Mol. Evol. 1998, 46, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Kranz, H.; Scholz, K.; Weisshaar, B. c-MYB oncogene-like genes encoding three MYB repeats occur in all major plant lineages. Plant J. 2000, 21, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Penfield, S.; Meissner, R.C.; Shoue, D.A.; Carpita, N.C.; Bevan, M.W. MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 2001, 13, 2777–2791. [Google Scholar] [CrossRef] [PubMed]
- Gigolashvili, T.; Yatusevich, R.; Berger, B.; Muller, C.; Flugge, U.I. The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007, 51, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Gigolashvili, T.; Engqvist, M.; Yatusevich, R.; Muller, C.; Flugge, U.I. HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana. New Phytol. 2008, 177, 627–642. [Google Scholar] [CrossRef] [PubMed]
- Gigolashvili, T.; Berger, B.; Flügge, U.I. Specific and coordinated control of indolic and aliphatic glucosinolate biosynthesis by R2R3-MYB transcription factors in Arabidopsis thaliana. Phytochem. Rev. 2009, 8, 3–13. [Google Scholar] [CrossRef]
- Takos, A.M.; Jaffe, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.P.; Walker, A.R. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007, 50, 660–677. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/MYB transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Feng, F.; Li, M.; Ma, F.; Cheng, L. Phenylpropanoid metabolites and expression of key genes involved in anthocyanin biosynthesis in the shaded peel of apple fruit in response to sun exposure. Plant Physiol. Biochem. 2013, 69, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Lai, L.B.; Nadeau, J.A.; Lucas, J.; Lee, E.K.; Nakagawa, T.; Zhao, L.; Geisler, M.; Sack, F.D. The Arabidopsis R2R3 MYB proteins FOUR LIPS and MYB88 restrict divisions late in the stomatal cell lineage. Plant Cell 2005, 17, 2754–2767. [Google Scholar] [CrossRef] [PubMed]
- Jakoby, M.J.; Falkenhan, D.; Mader, M.T.; Brininstool, G.; Wischnitzki, E.; Platz, N.; Hudson, A.; Hulskamp, M.; Larkin, J.; Schnittger, A. Transcriptional profiling of mature Arabidopsis trichomes reveals that NOECK encodes the MIXTA-like transcriptional regulator MYB106. Plant Physiol. 2008, 148, 1583–1602. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.H.; Kirik, V.; Hulskamp, M.; Nam, K.H.; Hagely, K.; Lee, M.M.; Schiefelbein, J. The MYB23 gene provides a positive feedback loop for cell fate specification in the Arabidopsis root epidermis. Plant Cell 2009, 21, 1080–1094. [Google Scholar] [CrossRef] [PubMed]
- Li, S.F.; Milliken, O.N.; Pham, H.; Seyit, R.; Napoli, R.; Preston, J.; Koltunow, A.M.; Parish, R.W. The Arabidopsis MYB5 transcription factor regulates mucilage synthesis, seed coat development, and trichome morphogenesis. Plant Cell 2009, 21, 72–89. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Lee, E.; Lucas, J.R.; Morohashi, K.; Li, D.; Murray, J.A.; Sack, F.D.; Grotewold, E. Regulation of cell proliferation in the stomatal lineage by the Arabidopsis MYB FOUR LIPS via direct targeting of core cell cycle genes. Plant Cell 2010, 22, 2306–2321. [Google Scholar] [CrossRef] [PubMed]
- Mengiste, T.; Chen, X.; Salmeron, J.; Dietrich, R. The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 2003, 15, 2551–2565. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.; Seo, J.S.; Han, S.W.; Koo, Y.J.; Kim, C.H.; Song, S.I.; Nahm, B.H.; Choi, Y.D.; Cheong, J.J. Over-expression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008, 146, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Devaiah, B.N.; Madhuvanthi, R.; Karthikeyan, A.S.; Raghothama, K.G. Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol. Plant 2009, 2, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Park, C.M. MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol. 2010, 186, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.A.; Gubler, F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 2005, 17, 705–721. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.B.; Zhu, J.; Gao, J.F.; Wang, C.; Li, H.; Li, H.; Zhang, H.Q.; Zhang, S.; Wang, D.M.; Wang, Q.X.; et al. Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. Plant J. 2007, 52, 528–538. [Google Scholar] [CrossRef] [PubMed]
- Shin, R.; Burch, A.Y.; Huppert, K.A.; Tiwari, S.B.; Murphy, A.S.; Guilfoyle, T.J.; Schachtman, D.P. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 2007, 19, 2440–2453. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Niu, Q.W.; Teng, C.; Li, C.; Mu, J.; Chua, N.H.; Zuo, J. Over-expression of PGA37/MYB118 and MYB115 promotes vegetative-to-embryonic transition in Arabidopsis. Cell Res. 2009, 19, 224–235. [Google Scholar] [CrossRef] [PubMed]
- Mu, R.L.; Cao, Y.R.; Liu, Y.F.; Lei, G.; Zou, H.F.; Liao, Y.; Wang, H.W.; Zhang, W.K.; Ma, B.; Du, J.Z.; et al. An R2R3-type transcription factor gene AtMYB59 regulates root growth and cell cycle progression in Arabidopsis. Cell Res. 2009, 19, 1291–1304. [Google Scholar] [CrossRef] [PubMed]
- Mandaokar, A. MYB108 acts together with MYB24 to regulate jasmonate-mediated stamen maturation in Arabidopsis. Plant Physiol. 2009, 149, 851–862. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.; Kim, J.H.; Nguyen, H.N.; Jikumaru, Y.; Kamiya, Y.; Hong, S.W.; Lee, H. A novel Arabidopsis MYB-like transcription factor, MYBH, regulates hypocotyl elongation by enhancing auxin accumulation. J. Exp. Bot. 2013, 64, 3911–3922. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chen, X.; Chen, J.; Xu, H.; Li, J.; Zhang, Z. Differential miRNA expression in Rehmannia glutinosa plants subjected to continuous cropping. BMC Plant Biol. 2011, 11, 53. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, Y.; Zhang, Y.; Chen, Y.; Bai, Y.; Wei, H.; Duan, H.; Zhou, C. Gene cloning, features of sequence, and analysis on temporal and spatial expression of Rehmannia glutinosa f. hueichingensis 3-ketoacyl CoA-thiolase. Chin. Tradit. Herbal Drugs 2013, 44, 76–84. [Google Scholar]
- Yang, Y.H.; Li, M.J.; Chen, X.J.; Wang, P.F.; Wang, F.Q.; Lin, W.X.; Yi, Y.J.; Zhang, Z.W.; Zhang, Z.Y. De novo characterization of the Rehmannia glutinosa leaf transcriptome and analysis of gene expression associated with replanting disease. Mol. Breed. 2014, 34, 905–915. [Google Scholar] [CrossRef]
- Wang, F.; Zhou, Y.; Huang, Y.; Li, M.; Tian, Y.; Feng, F.; Chen, X.; Zhang, Z. Cloning and expression analysis of the expansin gene RgEXPA10 in Rehmannia glutinosa. Acta Pharmacol. Sin. 2015, 50, 233–240. [Google Scholar]
- Yang, A.; Dai, X.; Zhang, W.H. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Yang, S.S.; Liang, Z.; Feng, B.R.; Liu, L.; Huang, Y.B.; Tang, Y.X. Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 2012, 12, 106. [Google Scholar] [CrossRef] [PubMed]
- Fornale, S.; Shi, X.; Chai, C.; Encina, A.; Irar, S.; Capellades, M.; Fuguet, E.; Torres, J.L.; Rovira, P.; Puigdomenech, P.; et al. ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. Plant J. 2010, 64, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; de Weese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Doerks, T.; Bork, P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015, 43, D257–D260. [Google Scholar] [CrossRef] [PubMed]
- Rose, A.; Meier, I.; Wienand, U. The tomato I-box binding factor LeMYBI is a member of a novel class of MYB-like proteins. Plant J. 1999, 20, 641–652. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, T.; Okamura, H.; Nagadoi, A.; Konig, P.; Rhodes, D.; Nishimura, Y. Solution structure of a telomeric DNA complex of human TRF1. Structure 2001, 9, 1237–1251. [Google Scholar] [CrossRef]
- Zhang, F.; Liu, X.; Zuo, K.; Sun, X.; Tang, K. Molecular cloning and expression analysis of a novel SANT/MYB gene from Gossypium barbadense. Mol. Biol. Rep. 2011, 38, 2329–2336. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Alabadı́, D.; Oyama, T.; Yanovsky, M.J.; Harmon, F.G.; Mas, P.; Kay, S.A. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 2001, 293, 880–883. [Google Scholar] [CrossRef] [PubMed]
- Bonke, M.; Thitamadee, S.; Mähönen, A.P.; Hauser, M.-T.; Helariutta, Y. APL regulates vascular tissue identity in Arabidopsis. Nature 2003, 426, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Smogorzewska, A.; van Steensel, B.; Bianchi, A.; Oelmann, S.; Schaefer, M.R.; Schnapp, G.; de Lange, T. Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 2000, 20, 1659–1668. [Google Scholar] [CrossRef] [PubMed]
- Karamysheva, Z.N.; Surovtseva, Y.V.; Vespa, L.; Shakirov, E.V.; Shippen, D.E. A C-terminal MYB extension domain defines a novel family of double-strand telomeric DNA-binding proteins in Arabidopsis. J. Biol. Chem. 2004, 279, 47799–47807. [Google Scholar] [CrossRef] [PubMed]
- Dvorackova, M.; Rossignol, P.; Shaw, P.J.; Koroleva, O.A.; Doonan, J.H.; Fajkus, J. AtTRB1, a telomeric DNA-binding protein from Arabidopsis, is concentrated in the nucleolus and shows highly dynamic association with chromatin. Plant J. 2010, 61, 637–649. [Google Scholar] [CrossRef] [PubMed]
- Yun, J.H.; Lee, W.K.; Kim, H.; Kim, E.; Cheong, C.; Cho, M.H.; Lee, W. Solution structure of telomere binding domain of AtTRB2 derived from Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2014, 452, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Illing, N.; Klak, C.; Johnson, C.; Brito, D.; Negrao, N.; Baine, F.; van Kets, V.; Ramchurn, K.R.; Seoighe, C.; Roden, L. Duplication of the Asymmetric Leaves1/Rough Sheath 2/Phantastica (ARP) gene precedes the explosive radiation of the Ruschioideae. Dev. Genes Evol. 2009, 219, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Haga, N.; Kato, K.; Murase, M.; Araki, S.; Kubo, M.; Demura, T.; Suzuki, K.; Müller, I.; Voß, U.; Jürgens, G. R1R2R3-Myb proteins positively regulate cytokinesis through activation of KNOLLE transcription in Arabidopsis thaliana. Development 2007, 134, 1101–1110. [Google Scholar] [CrossRef] [PubMed]
- Saeed, A.I.; Bhagabati, N.K.; Braisted, J.C.; Liang, W.; Sharov, V.; Howe, E.A.; Li, J.; Thiagarajan, M.; White, J.A.; Quackenbush, J. TM4 microarray software suite. Methods Enzymol. 2006, 411, 134–193. [Google Scholar] [PubMed]
- Romero, I.; Fuertes, A.; Benito, M.J.; Malpica, J.M.; Leyva, A.; Paz-Ares, J. More than 80 R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J. 1998, 14, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Katiyar, A.; Smita, S.; Lenka, S.K.; Rajwanshi, R.; Chinnusamy, V.; Bansal, K.C. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics 2012, 13, 544. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Holtgrawe, D.; Schneider, J.; Pucker, B.; Sorensen, T.R.; Weisshaar, B. Genome-wide identification and characterisation of R2R3-MYB genes in sugar beet (Beta vulgaris). BMC Plant Biol. 2014, 14, 249. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Feng, B.R.; Yang, S.S.; Huang, Y.B.; Tang, Y.X. The R2R3-MYB transcription factor gene family in maize. PLoS ONE 2012, 7, e37463. [Google Scholar] [CrossRef] [PubMed]
- Yanhui, C.; Xiaoyuan, Y.; Kun, H.; Meihua, L.; Jigang, L.; Zhaofeng, G.; Zhiqiang, L.; Yunfei, Z.; Xiaoxiao, W.; Xiaoming, Q.; et al. The MYB transcription factor superfamily of Arabidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 2006, 60, 107–124. [Google Scholar] [CrossRef] [PubMed]
- Matus, J.T.; Aquea, F.; Arce-Johnson, P. Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol. 2008, 8, 83. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, O.; Nahal, H.; Foong, J.; Provart, N.J.; Campbell, M.M. Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 2009, 149, 981–993. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.H.; Zhang, S.Z.; Wang, R.K.; Zhang, R.F.; Hao, Y.J. Genome wide analysis of the apple MYB transcription factor family allows the identification of MdoMYB121 gene confering abiotic stress tolerance in plants. PLoS ONE 2013, 8, e69955. [Google Scholar] [CrossRef] [PubMed]
- Frith, M.C.; Saunders, N.F.; Kobe, B.; Bailey, T.L. Discovering sequence motifs with arbitrary insertions and deletions. PLoS Comput. Biol. 2008, 4, e1000071. [Google Scholar] [CrossRef] [PubMed]
- Ogata, K.; Morikawa, S.; Nakamura, H.; Sekikawa, A.; Inoue, T.; Kanai, H.; Sarai, A.; Ishii, S.; Nishimura, Y. Solution structure of a specific DNA complex of the MYB DNA-binding domain with cooperative recognition helices. Cell 1994, 79, 639–648. [Google Scholar] [CrossRef]
- Ogata, K.; Hojo, H.; Aimoto, S.; Nakai, T.; Nakamura, H.; Sarai, A.; Ishii, S.; Nishimura, Y. Solution structure of a DNA-binding unit of MYB: A helix-turn-helix-related motif with conserved tryptophans forming a hydrophobic core. Proc. Natl. Acad. Sci. USA 1992, 89, 6428–6432. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Gu, X.; Peterson, T. Identification of conserved gene structures and carboxy-terminal motifs in the MYB gene family of Arabidopsis and Oryza sativa L. ssp. indica. Genome Biol. 2004, 5, R46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waites, R.; Selvadurai, H.R.; Oliver, I.R.; Hudson, A. The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 1998, 93, 779–789. [Google Scholar] [CrossRef]
- Tsiantis, M.; Schneeberger, R.; Golz, J.F.; Freeling, M.; Langdale, J.A. The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Science 1999, 284, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Jiang, C.; Xing, J.; Dong, J. Structure and function of the 22nd subfamily in Arabidopsis R2R3-MYB family. Yi Chuan 2014, 36, 985–994. [Google Scholar] [PubMed]
- Cheong, Y.H.; Chang, H.S.; Gupta, R.; Wang, X.; Zhu, T.; Luan, S. Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 2002, 129, 661–677. [Google Scholar] [CrossRef] [PubMed]
- Kamei, A.; Seki, M.; Umezawa, T.; Ishida, J.; Satou, M.; Akiyama, K.; Zhu, J.; Shinozaki, K. Analysis of gene expression profiles in Arabidopsis salt overly sensitive mutants sos2-1 and sos3-1. Plant Cell Environ. 2005, 28, 1267–1275. [Google Scholar] [CrossRef]
- Agarwal, M.; Hao, Y.; Kapoor, A.; Dong, C.H.; Fujii, H.; Zheng, X.; Zhu, J.K. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 2006, 281, 37636–37645. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yu, C.; Li, Y.; Lam, T.W.; Yiu, S.M.; Kristiansen, K.; Wang, J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 2009, 25, 1966–1967. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, D.L.; Chappey, C.; Lash, A.E.; Leipe, D.D.; Madden, T.L.; Schuler, G.D.; Tatusova, T.A.; Rapp, B.A. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2000, 28, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative Ct method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, F.; Suo, Y.; Wei, H.; Li, M.; Xie, C.; Wang, L.; Chen, X.; Zhang, Z. Identification and Characterization of 40 Isolated Rehmannia glutinosa MYB Family Genes and Their Expression Profiles in Response to Shading and Continuous Cropping. Int. J. Mol. Sci. 2015, 16, 15009-15030. https://doi.org/10.3390/ijms160715009
Wang F, Suo Y, Wei H, Li M, Xie C, Wang L, Chen X, Zhang Z. Identification and Characterization of 40 Isolated Rehmannia glutinosa MYB Family Genes and Their Expression Profiles in Response to Shading and Continuous Cropping. International Journal of Molecular Sciences. 2015; 16(7):15009-15030. https://doi.org/10.3390/ijms160715009
Chicago/Turabian StyleWang, Fengqing, Yanfei Suo, He Wei, Mingjie Li, Caixia Xie, Lina Wang, Xinjian Chen, and Zhongyi Zhang. 2015. "Identification and Characterization of 40 Isolated Rehmannia glutinosa MYB Family Genes and Their Expression Profiles in Response to Shading and Continuous Cropping" International Journal of Molecular Sciences 16, no. 7: 15009-15030. https://doi.org/10.3390/ijms160715009