NKG2A/CD94 Is a New Immune Receptor for HLA-G and Distinguishes Amino Acid Differences in the HLA-G Heavy Chain
Abstract
:1. Introduction
2. Results
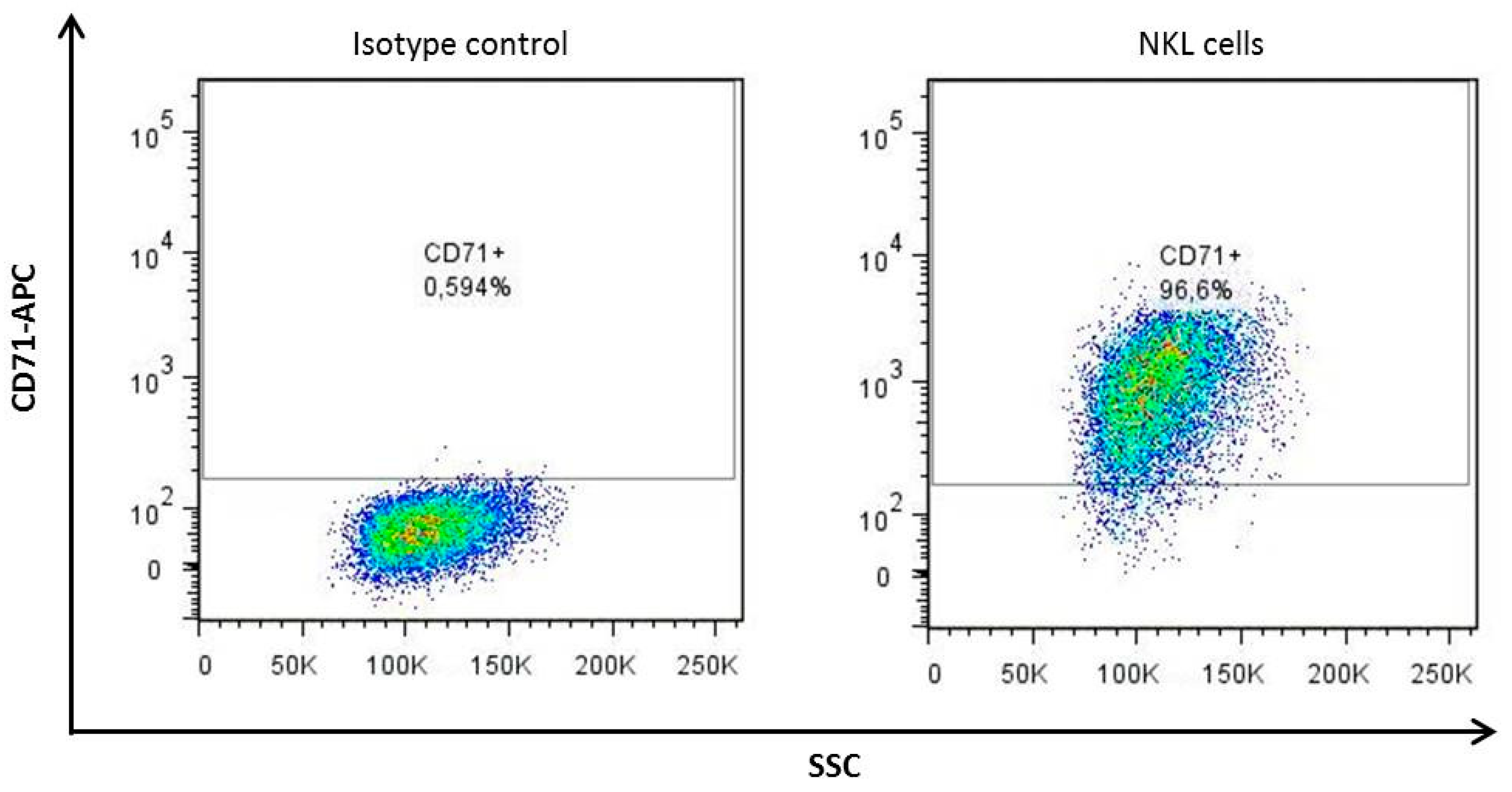
2.1. The TriCEPS Method Can Be Performed Using NKL Cells as Target Cells and sHLA-G*01:01-TriCEPS as Ligand
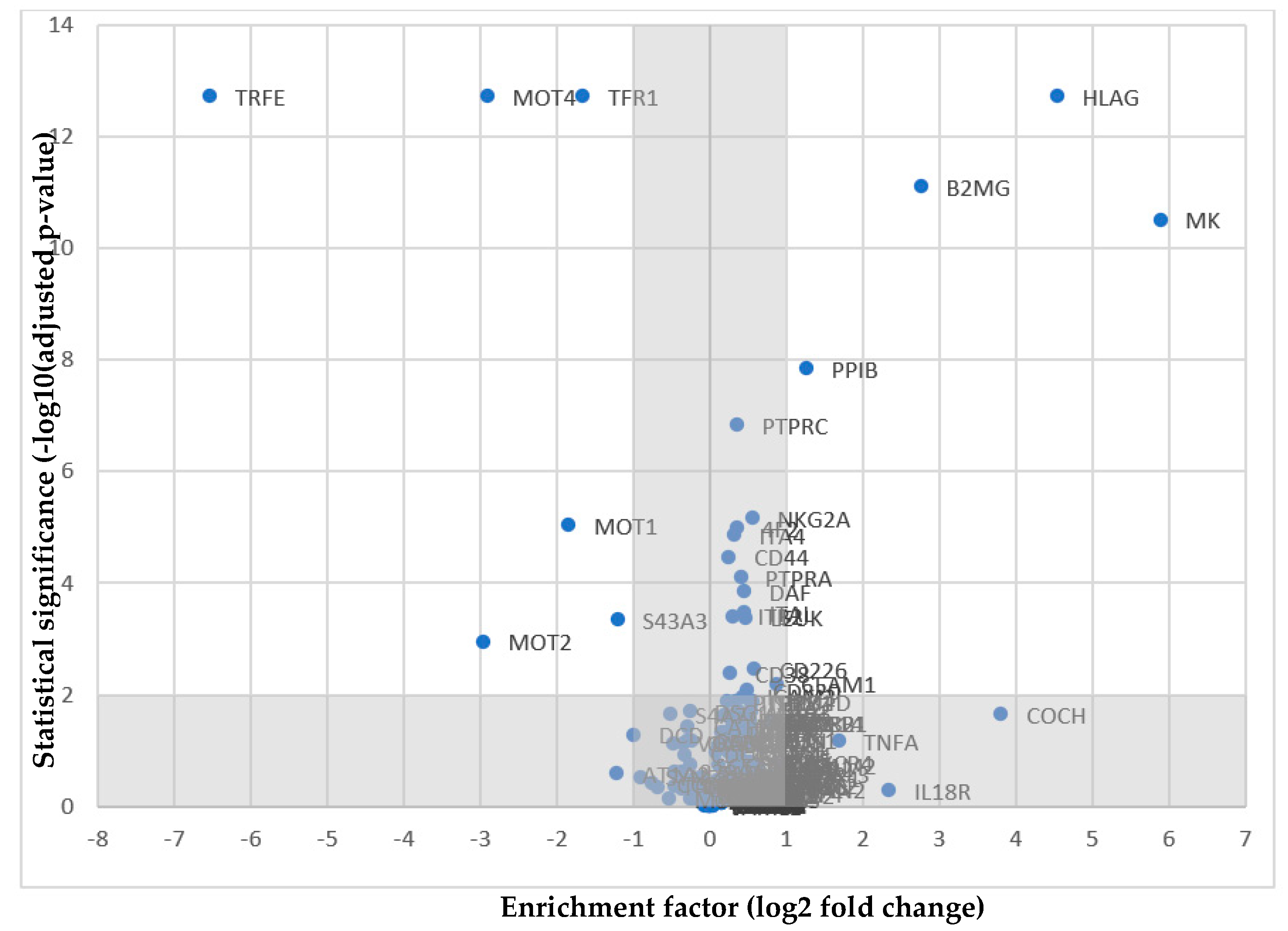
2.2. NKG2A as a Potential Receptor for HLA-G
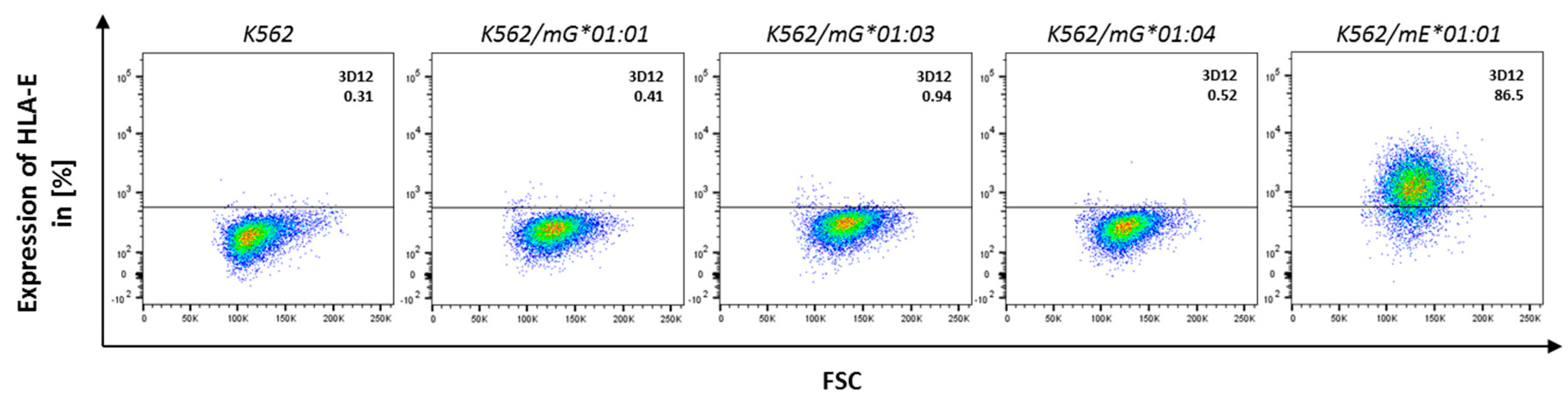
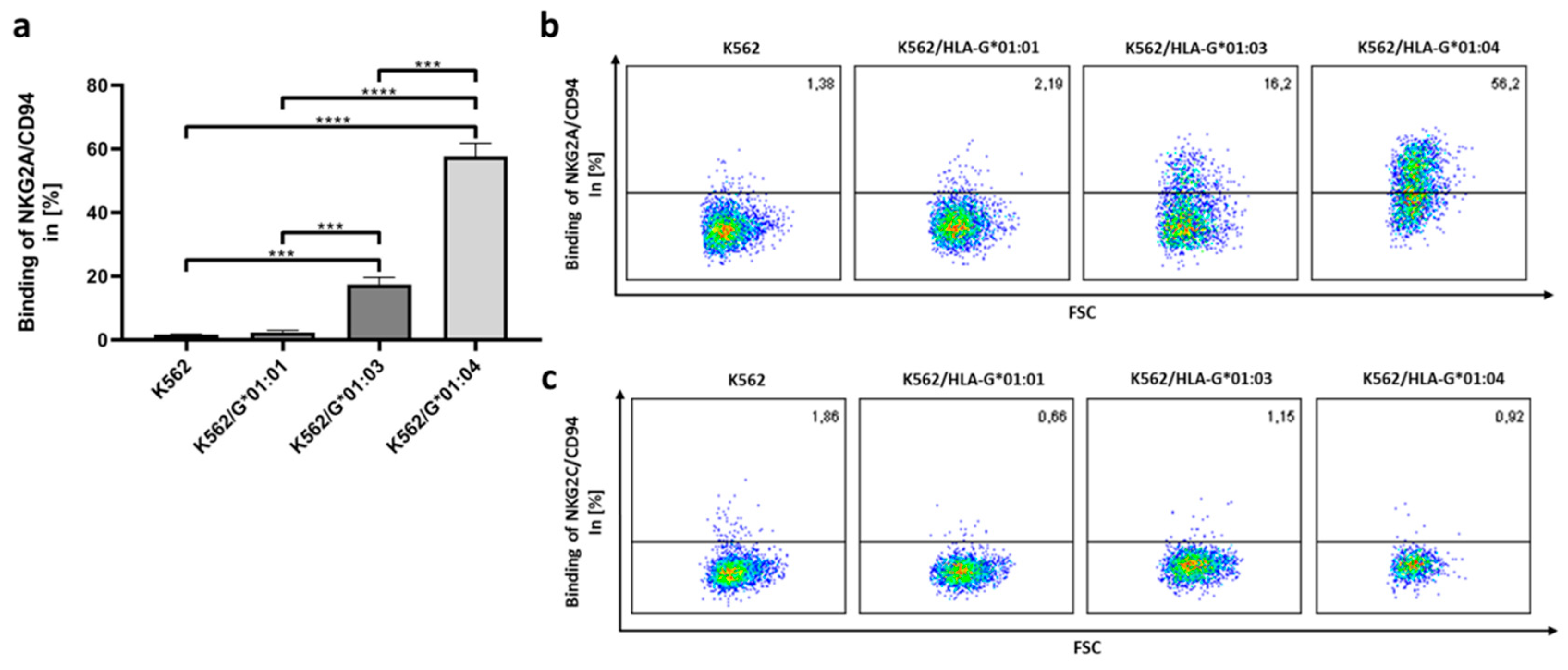
2.3. NKG2A/CD94 Distinguishes AA Differences in the HLA-G Heavy Chain
3. Discussion
4. Materials and Methods
4.1. Maintenance of the Cell Lines
4.2. Cloning of HLA-G Constructs
4.3. Cloning of Plasmid Encoding for Soluble NKG2/CD94 Heterodimers
4.4. Stable Lentiviral Transduction of K562 Cells with HLA-G and NKG2/CD94 Constructs
4.5. Large-Scale Production of sHLA-G, sNKG2A, and sNKG2C
4.6. LCR-TriCEPS Method for Capturing of Receptors for HLA-G
4.7. Detection of sNKG2A/CD94 Binding to HLA-G
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AA | amino acid |
| ß2m | β2-microglobulin |
| DC | dendritic cell |
| HLA | human leukocyte antigen |
| HSCT | hematopoietic stem cell transplantation |
| ITIMs | immune receptor tyrosine-based inhibitory motifs |
| NK cell | natural killer cell |
| mHLA-G | membrane-bound HLA-G |
| sHLA-G | soluble HLA-G |
| sNKG2A/CD94 | soluble NKG2A/CD94 |
| sNKG2C/CD94 | soluble NKG2C/CD94 |
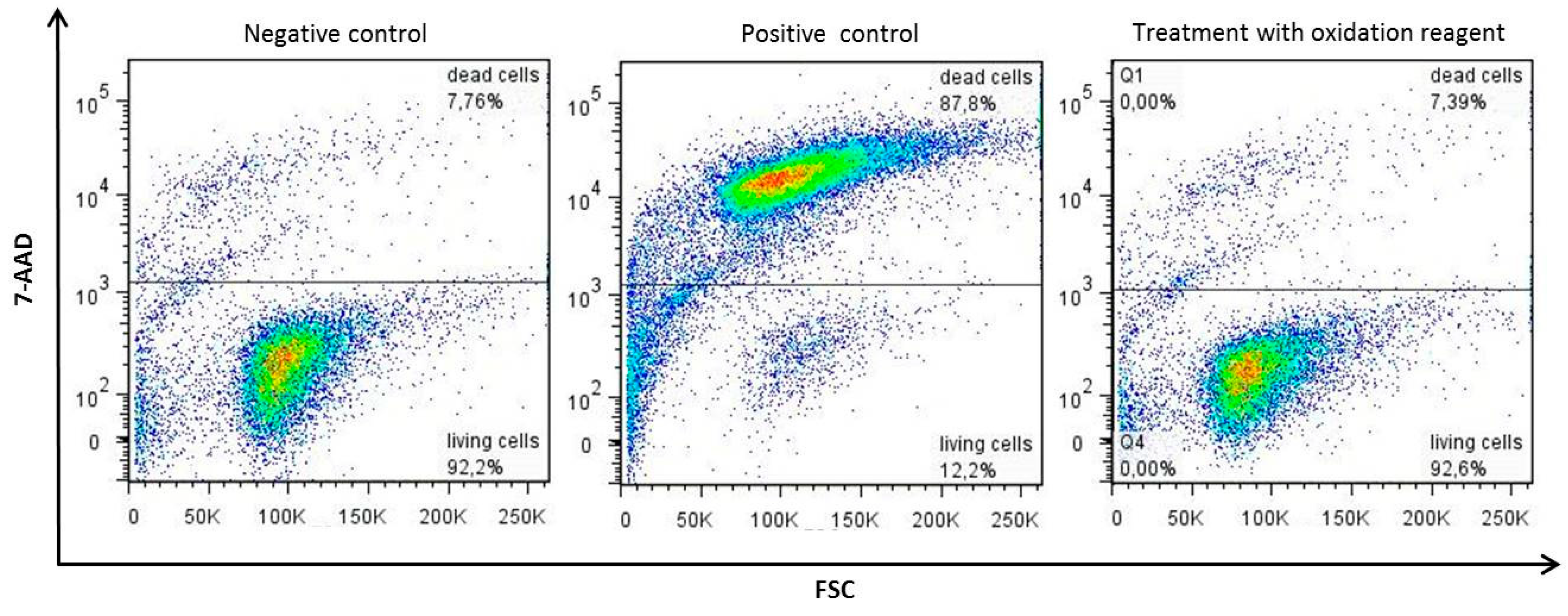
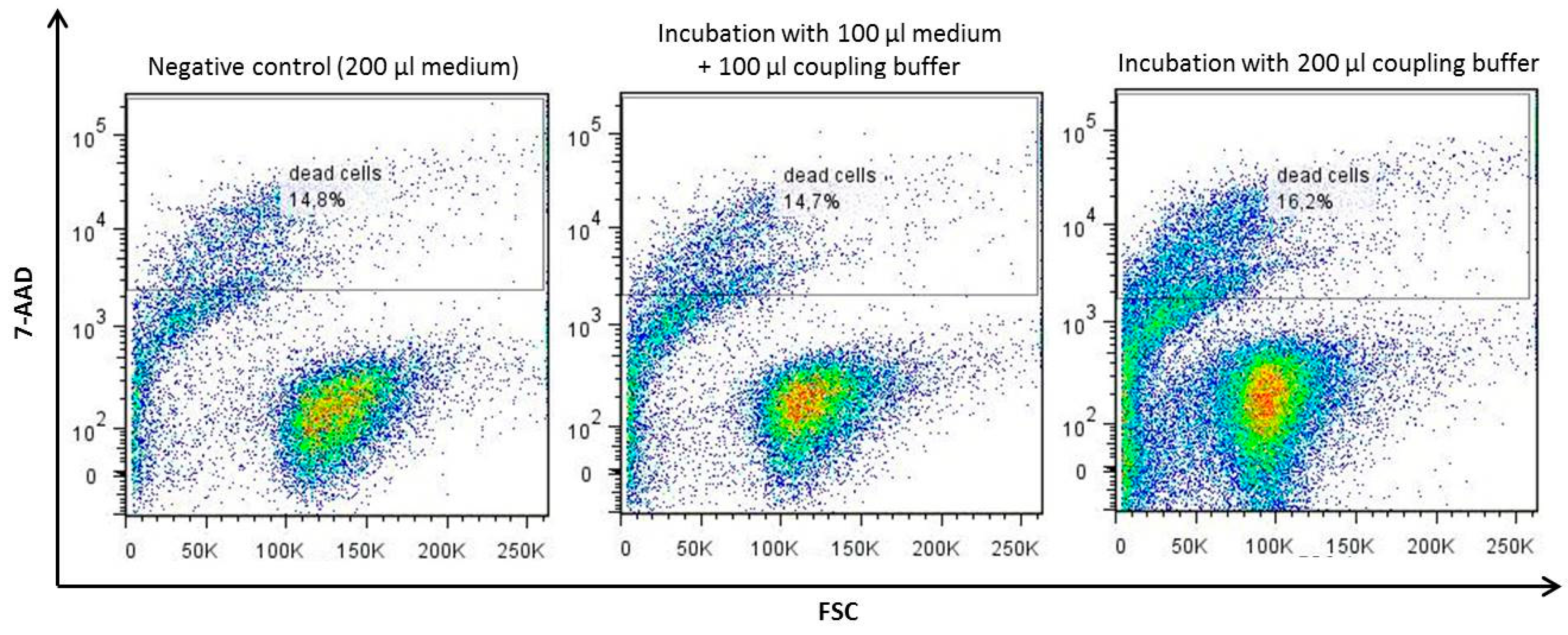
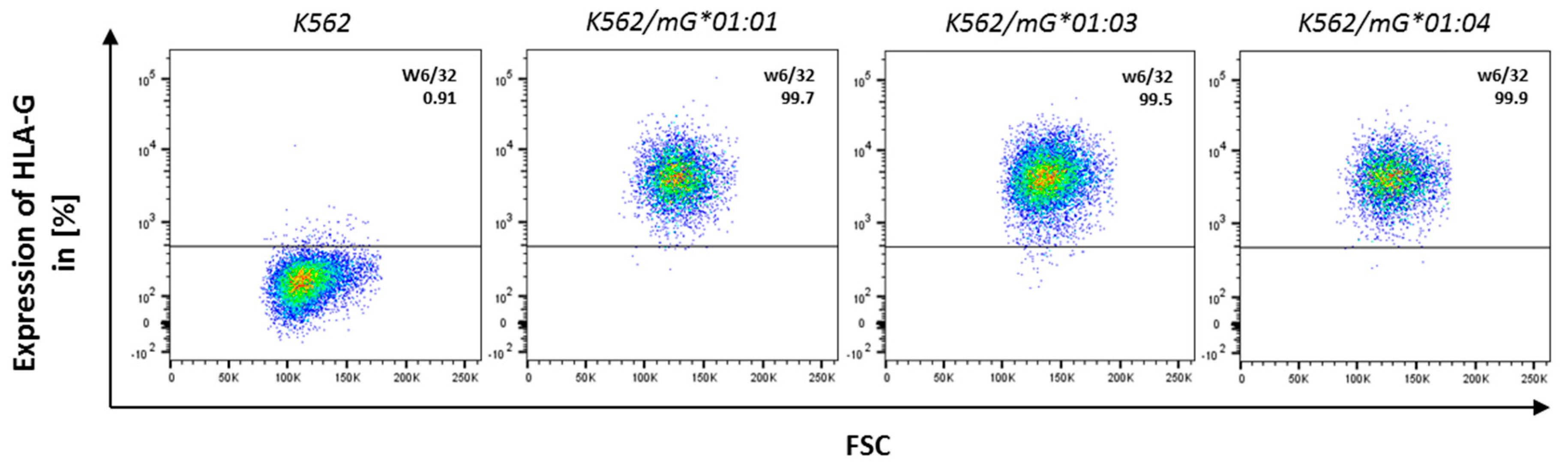
Appendix A


Native PAGE for Verification of Heterodimeric Complex of NKG2A/CD94 and NKG2C/CD94

References
- Kovats, S.; Main, E.K.; Librach, C.; Stubblebine, M.; Fisher, S.J.; DeMars, R. A class I antigen, HLA-G, expressed in human trophoblasts. Science 1990, 248, 220–223. [Google Scholar] [CrossRef] [PubMed]
- Hunt, J.S.; Petroff, M.G.; McIntire, R.H.; Ober, C. HLA-G and immune tolerance in pregnancy. FASEB J. 2005, 19, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Yie, S.M.; Li, L.H.; Li, Y.M.; Librach, C. HLA-G protein concentrations in maternal serum and placental tissue are decreased in preeclampsia. Am. J. Obstet. Gynecol. 2004, 191, 525–529. [Google Scholar] [CrossRef] [PubMed]
- Colbern, G.T.; Chiang, M.H.; Main, E.K. Expression of the nonclassic histocompatibility antigen HLA-G by preeclamptic placenta. Am. J. Obstet. Gynecol. 1994, 170, 1244–1250. [Google Scholar] [CrossRef]
- Peng, B.; Zhang, L.; Xing, A.Y.; Hu, M.; Liu, S.Y. The expression of human leukocyte antigen G and E on human first trimester placenta and its relationship with recurrent spontaneous abortion. J. Sichuan Univ. 2008, 39, 976–979. [Google Scholar]
- Le Discorde, M.; Moreau, P.; Sabatier, P.; Legeais, J.M.; Carosella, E.D. Expression of HLA-G in human cornea, an immune-privileged tissue. Hum. Immunol. 2003, 64, 1039–1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mallet, V.; Blaschitz, A.; Crisa, L.; Schmitt, C.; Fournel, S.; King, A.; Loke, Y.W.; Dohr, G.; Le Bouteiller, P. HLA-G in the human thymus: A subpopulation of medullary epithelial but not CD83+ dendritic cells expresses HLA-G as a membrane-bound and soluble protein. Int. Immunol. 1999, 11, 889–898. [Google Scholar] [CrossRef] [Green Version]
- Shukla, H.; Swaroop, A.; Srivastava, R.; Weissman, S.M. The mRNA of a human class I gene HLA G/HLA 6.0 exhibits a restricted pattern of expression. Nucleic Acids Res. 1990, 18, 2189. [Google Scholar] [CrossRef] [Green Version]
- Blaschitz, A.; Lenfant, F.; Mallet, V.; Hartmann, M.; Bensussan, A.; Geraghty, D.E.; Le Bouteiller, P.; Dohr, G. Endothelial cells in chorionic fetal vessels of first trimester placenta express HLA-G. Eur. J. Immunol. 1997, 27, 3380–3388. [Google Scholar] [CrossRef]
- Rizzo, R.; Andersen, A.S.; Lassen, M.R.; Sorensen, H.C.; Bergholt, T.; Larsen, M.H.; Melchiorri, L.; Stignani, M.; Baricordi, O.R.; Hviid, T.V. Soluble human leukocyte antigen-G isoforms in maternal plasma in early and late pregnancy. Am. J. Reprod. Immunol. 2009, 62, 320–338. [Google Scholar] [CrossRef]
- Rebmann, V.; Busemann, A.; Lindemann, M.; Grosse-Wilde, H. Detection of HLA-G5 secreting cells. Hum. Immunol. 2003, 64, 1017–1024. [Google Scholar] [CrossRef] [PubMed]
- Donaghy, L.; Gros, F.; Amiot, L.; Mary, C.; Maillard, A.; Guiguen, C.; Gangneux, J.P. Elevated levels of soluble non-classical major histocompatibility class I molecule human leucocyte antigen (HLA)-G in the blood of HIV-infected patients with or without visceral leishmaniasis. Clin. Exp. Immunol. 2007, 147, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Wiendl, H.; Mitsdoerffer, M.; Hofmeister, V.; Wischhusen, J.; Bornemann, A.; Meyermann, R.; Weiss, E.H.; Melms, A.; Weller, M. A functional role of HLA-G expression in human gliomas: An alternative strategy of immune escape. J. Immunol. 2002, 168, 4772–4780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rebmann, V.; Regel, J.; Stolke, D.; Grosse-Wilde, H. Secretion of sHLA-G molecules in malignancies. Semin. Cancer Biol. 2003, 13, 371–377. [Google Scholar] [CrossRef]
- Ugurel, S.; Rebmann, V.; Ferrone, S.; Tilgen, W.; Grosse-Wilde, H.; Reinhold, U. Soluble human leukocyte antigen—G serum level is elevated in melanoma patients and is further increased by interferon-alpha immunotherapy. Cancer 2001, 92, 369–376. [Google Scholar] [CrossRef]
- Gros, F.; Sebti, Y.; de Guibert, S.; Branger, B.; Bernard, M.; Fauchet, R.; Amiot, L. Soluble HLA-G molecules increase during acute leukemia, especially in subtypes affecting monocytic and lymphoid lineages. Neoplasia 2006, 8, 223–230. [Google Scholar] [CrossRef]
- Verbruggen, L.A.; Rebmann, V.; Demanet, C.; De Cock, S.; Grosse-Wilde, H. Soluble HLA-G in rheumatoid arthritis. Hum. Immunol. 2006, 67, 561–567. [Google Scholar] [CrossRef]
- Fainardi, E.; Rizzo, R.; Melchiorri, L.; Vaghi, L.; Castellazzi, M.; Marzola, A.; Govoni, V.; Paolino, E.; Tola, M.R.; Granieri, E.; et al. Presence of detectable levels of soluble HLA-G molecules in CSF of relapsing-remitting multiple sclerosis: Relationship with CSF soluble HLA-I and IL-10 concentrations and MRI findings. J. Neuroimmunol. 2003, 142, 149–158. [Google Scholar] [CrossRef]
- Crispim, J.C.; Duarte, R.A.; Soares, C.P.; Costa, R.; Silva, J.S.; Mendes-Junior, C.T.; Wastowski, I.J.; Faggioni, L.P.; Saber, L.T.; Donadi, E.A. Human leukocyte antigen-G expression after kidney transplantation is associated with a reduced incidence of rejection. Transpl. Immunol. 2008, 18, 361–367. [Google Scholar] [CrossRef]
- Robinson, J.; Halliwell, J.A.; Hayhurst, J.D.; Flicek, P.; Parham, P.; Marsh, S.G. The IPD and IMGT/HLA database: Allele variant databases. Nucleic Acids Res. 2015, 43, D423–D431. [Google Scholar] [CrossRef] [Green Version]
- Diehl, M.; Munz, C.; Keilholz, W.; Stevanovic, S.; Holmes, N.; Loke, Y.W.; Rammensee, H.G. Nonclassical HLA-G molecules are classical peptide presenters. Curr. Biol. 1996, 6, 305–314. [Google Scholar] [CrossRef] [Green Version]
- Romagnani, C.; Pietra, G.; Falco, M.; Millo, E.; Mazzarino, P.; Biassoni, R.; Moretta, A.; Moretta, L.; Mingari, M.C. Identification of HLA-E-specific alloreactive T lymphocytes: A cell subset that undergoes preferential expansion in mixed lymphocyte culture and displays a broad cytolytic activity against allogeneic cells. Proc. Natl. Acad. Sci. USA 2002, 99, 11328–11333. [Google Scholar] [CrossRef] [Green Version]
- Kraemer, T.; Celik, A.A.; Huyton, T.; Kunze-Schumacher, H.; Blasczyk, R.; Bade-Doding, C. HLA-E: Presentation of a Broader Peptide Repertoire Impacts the Cellular Immune Response-Implications on HSCT Outcome. Stem Cells Int. 2015, 2015, 346714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Celik, A.A.; Simper, G.; Huyton, T.; Blasczyk, R.; Bade-Doeding, C. The Bound Peptides Are the Determinants for the Potential of Hla-G Mediated Immune Regulation. Hum. Immunol. 2017, 78, 1. [Google Scholar] [CrossRef]
- Matte, C.; Lacaille, J.; Zijenah, L.; Ward, B.; Roger, M.; The ZVITAMBO Study Group. HLA-G and HLA-E polymorphisms in an indigenous African population. Hum. Immunol. 2000, 61, 1150–1156. [Google Scholar] [CrossRef]
- Castelli, E.C.; Ramalho, J.; Porto, I.O.; Lima, T.H.; Felicio, L.P.; Sabbagh, A.; Donadi, E.A.; Mendes-Junior, C.T. Insights into HLA-G Genetics Provided by Worldwide Haplotype Diversity. Front. Immunol. 2014, 5, 476. [Google Scholar] [CrossRef] [Green Version]
- Van der Ven, K.; Skrablin, S.; Engels, G.; Krebs, D. HLA-G polymorphisms and allele frequencies in Caucasians. Hum. Immunol. 1998, 59, 302–312. [Google Scholar] [CrossRef]
- Carosella, E.D.; Rouas-Freiss, N.; Tronik-Le Roux, D.; Moreau, P.; LeMaoult, J. HLA-G: An Immune Checkpoint Molecule. Adv. Immunol. 2015, 127, 33–144. [Google Scholar] [CrossRef]
- Alegre, E.; Rizzo, R.; Bortolotti, D.; Fernandez-Landazuri, S.; Fainardi, E.; Gonzalez, A. Some basic aspects of HLA-G biology. J. Immunol. Res. 2014, 2014, 657625. [Google Scholar] [CrossRef] [Green Version]
- Rouas-Freiss, N.; Moreau, P.; Menier, C.; LeMaoult, J.; Carosella, E.D. Expression of tolerogenic HLA-G molecules in cancer prevents antitumor responses. Semin. Cancer Biol. 2007, 17, 413–421. [Google Scholar] [CrossRef]
- Rebmann, V.; da Silva Nardi, F.; Wagner, B.; Horn, P.A. HLA-G as a tolerogenic molecule in transplantation and pregnancy. J. Immunol. Res. 2014, 2014, 297073. [Google Scholar] [CrossRef] [PubMed]
- Cosman, D.; Fanger, N.; Borges, L.; Kubin, M.; Chin, W.; Peterson, L.; Hsu, M.L. A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules. Immunity 1997, 7, 273–282. [Google Scholar] [CrossRef] [Green Version]
- Colonna, M.; Navarro, F.; Bellon, T.; Llano, M.; Garcia, P.; Samaridis, J.; Angman, L.; Cella, M.; Lopez-Botet, M. A common inhibitory receptor for major histocompatibility complex class I molecules on human lymphoid and myelomonocytic cells. J. Exp. Med. 1997, 186, 1809–1818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, A.; Rebmann, V.; LeMaoult, J.; Horn, P.A.; Carosella, E.D.; Alegre, E. The immunosuppressive molecule HLA-G and its clinical implications. Crit. Rev. Clin. Lab. Sci. 2012, 49, 63–84. [Google Scholar] [CrossRef] [PubMed]
- Shiroishi, M.; Tsumoto, K.; Amano, K.; Shirakihara, Y.; Colonna, M.; Braud, V.M.; Allan, D.S.; Makadzange, A.; Rowland-Jones, S.; Willcox, B.; et al. Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc. Natl. Acad. Sci. USA 2003, 100, 8856–8861. [Google Scholar] [CrossRef] [Green Version]
- Rajagopalan, S.; Long, E.O. A human histocompatibility leukocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells. J. Exp. Med. 1999, 189, 1093–1100. [Google Scholar] [CrossRef] [Green Version]
- Contini, P.; Ghio, M.; Poggi, A.; Filaci, G.; Indiveri, F.; Ferrone, S.; Puppo, F. Soluble HLA-A,-B,-C and -G molecules induce apoptosis in T and NK CD8+ cells and inhibit cytotoxic T cell activity through CD8 ligation. Eur. J. Immunol. 2003, 33, 125–134. [Google Scholar] [CrossRef]
- LeMaoult, J.; Zafaranloo, K.; Le Danff, C.; Carosella, E.D. HLA-G up-regulates ILT2, ILT3, ILT4, and KIR2DL4 in antigen presenting cells, NK cells, and T cells. FASEB J. 2005, 19, 662–664. [Google Scholar] [CrossRef]
- Fons, P.; Chabot, S.; Cartwright, J.E.; Lenfant, F.; L’Faqihi, F.; Giustiniani, J.; Herault, J.P.; Gueguen, G.; Bono, F.; Savi, P.; et al. Soluble HLA-G1 inhibits angiogenesis through an apoptotic pathway and by direct binding to CD160 receptor expressed by endothelial cells. Blood 2006, 108, 2608–2615. [Google Scholar] [CrossRef] [Green Version]
- Kapasi, K.; Albert, S.E.; Yie, S.; Zavazava, N.; Librach, C.L. HLA-G has a concentration-dependent effect on the generation of an allo-CTL response. Immunology 2000, 101, 191–200. [Google Scholar] [CrossRef]
- Liang, S.; Ristich, V.; Arase, H.; Dausset, J.; Carosella, E.D.; Horuzsko, A. Modulation of dendritic cell differentiation by HLA-G and ILT4 requires the IL-6--STAT3 signaling pathway. Proc. Natl. Acad. Sci. USA 2008, 105, 8357–8362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregori, S.; Tomasoni, D.; Pacciani, V.; Scirpoli, M.; Battaglia, M.; Magnani, C.F.; Hauben, E.; Roncarolo, M.G. Differentiation of type 1 T regulatory cells (Tr1) by tolerogenic DC-10 requires the IL-10-dependent ILT4/HLA-G pathway. Blood 2010, 116, 935–944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrie, E.J.; Clements, C.S.; Lin, J.; Sullivan, L.C.; Johnson, D.; Huyton, T.; Heroux, A.; Hoare, H.L.; Beddoe, T.; Reid, H.H.; et al. CD94-NKG2A recognition of human leukocyte antigen (HLA)-E bound to an HLA class I leader sequence. J. Exp. Med. 2008, 205, 725–735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoare, H.L.; Sullivan, L.C.; Clements, C.S.; Ely, L.K.; Beddoe, T.; Henderson, K.N.; Lin, J.; Reid, H.H.; Brooks, A.G.; Rossjohn, J. Subtle changes in peptide conformation profoundly affect recognition of the non-classical MHC class I molecule HLA-E by the CD94-NKG2 natural killer cell receptors. J. Mol. Biol. 2008, 377, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Vales-Gomez, M.; Reyburn, H.T.; Erskine, R.A.; Lopez-Botet, M.; Strominger, J.L. Kinetics and peptide dependency of the binding of the inhibitory NK receptor CD94/NKG2-A and the activating receptor CD94/NKG2-C to HLA-E. EMBO J. 1999, 18, 4250–4260. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, B.K.; Barahmand-Pour, F.; Paulsene, W.; Medley, S.; Geraghty, D.E.; Strong, R.K. Interactions between NKG2x immunoreceptors and HLA-E ligands display overlapping affinities and thermodynamics. J. Immunol. 2005, 174, 2878–2884. [Google Scholar] [CrossRef] [Green Version]
- Perez-Villar, J.J.; Melero, I.; Navarro, F.; Carretero, M.; Bellon, T.; Llano, M.; Colonna, M.; Geraghty, D.E.; Lopez-Botet, M. The CD94/NKG2-A inhibitory receptor complex is involved in natural killer cell-mediated recognition of cells expressing HLA-G1. J. Immunol. 1997, 158, 5736–5743. [Google Scholar]
- Soderstrom, K.; Corliss, B.; Lanier, L.L.; Phillips, J.H. CD94/NKG2 is the predominant inhibitory receptor involved in recognition of HLA-G by decidual and peripheral blood NK cells. J. Immunol. 1997, 159, 1072–1075. [Google Scholar]
- Navarro, F.; Llano, M.; Bellon, T.; Colonna, M.; Geraghty, D.E.; Lopez-Botet, M. The ILT2(LIR1) and CD94/NKG2A NK cell receptors respectively recognize HLA-G1 and HLA-E molecules co-expressed on target cells. Eur. J. Immunol. 1999, 29, 277–283. [Google Scholar] [CrossRef]
- Schmidt, S.; Tramsen, L.; Rais, B.; Ullrich, E.; Lehrnbecher, T. Natural killer cells as a therapeutic tool for infectious diseases—Current status and future perspectives. Oncotarget 2018, 9, 20891–20907. [Google Scholar] [CrossRef] [Green Version]
- Celik, A.A.; Simper, G.S.; Hiemisch, W.; Blasczyk, R.; Bade-Doding, C. HLA-G peptide preferences change in transformed cells: Impact on the binding motif. Immunogenetics 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuroki, K.; Matsubara, H.; Kanda, R.; Miyashita, N.; Shiroishi, M.; Fukunaga, Y.; Kamishikiryo, J.; Fukunaga, A.; Fukuhara, H.; Hirose, K.; et al. Structural and Functional Basis for LILRB Immune Checkpoint Receptor Recognition of HLA-G Isoforms. J. Immunol. 2019, 203, 3386–3394. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Song, H.; Cheng, H.; Qi, J.; Nam, G.; Tan, S.; Wang, J.; Fang, M.; Shi, Y.; Tian, Z.; et al. Structures of the four Ig-like domain LILRB2 and the four-domain LILRB1 and HLA-G1 complex. Cell Mol. Immunol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Chapman, T.L.; Heikeman, A.P.; Bjorkman, P.J. The inhibitory receptor LIR-1 uses a common binding interaction to recognize class I MHC molecules and the viral homolog UL18. Immunity 1999, 11, 603–613. [Google Scholar] [CrossRef] [Green Version]
- Allan, D.S.; Colonna, M.; Lanier, L.L.; Churakova, T.D.; Abrams, J.S.; Ellis, S.A.; McMichael, A.J.; Braud, V.M. Tetrameric complexes of human histocompatibility leukocyte antigen (HLA)-G bind to peripheral blood myelomonocytic cells. J. Exp. Med. 1999, 189, 1149–1156. [Google Scholar] [CrossRef]
- Shiroishi, M.; Kuroki, K.; Rasubala, L.; Tsumoto, K.; Kumagai, I.; Kurimoto, E.; Kato, K.; Kohda, D.; Maenaka, K. Structural basis for recognition of the nonclassical MHC molecule HLA-G by the leukocyte Ig-like receptor B2 (LILRB2/LIR2/ILT4/CD85d). Proc. Natl. Acad. Sci. USA 2006, 103, 16412–16417. [Google Scholar] [CrossRef] [Green Version]
- Colonna, M.; Samaridis, J.; Cella, M.; Angman, L.; Allen, R.L.; O’Callaghan, C.A.; Dunbar, R.; Ogg, G.S.; Cerundolo, V.; Rolink, A. Human myelomonocytic cells express an inhibitory receptor for classical and nonclassical MHC class I molecules. J. Immunol. 1998, 160, 3096–3100. [Google Scholar]
- Ponte, M.; Cantoni, C.; Biassoni, R.; Tradori-Cappai, A.; Bentivoglio, G.; Vitale, C.; Bertone, S.; Moretta, A.; Moretta, L.; Mingari, M.C. Inhibitory receptors sensing HLA-G1 molecules in pregnancy: Decidua-associated natural killer cells express LIR-1 and CD94/NKG2A and acquire p49, an HLA-G1-specific receptor. Proc. Natl. Acad. Sci. USA 1999, 96, 5674–5679. [Google Scholar] [CrossRef] [Green Version]
- Cantoni, C.; Verdiani, S.; Falco, M.; Pessino, A.; Cilli, M.; Conte, R.; Pende, D.; Ponte, M.; Mikaelsson, M.S.; Moretta, L.; et al. p49, a putative HLA class I-specific inhibitory NK receptor belonging to the immunoglobulin superfamily. Eur. J. Immunol. 1998, 28, 1980–1990. [Google Scholar] [CrossRef]
- Fournel, S.; Aguerre-Girr, M.; Huc, X.; Lenfant, F.; Alam, A.; Toubert, A.; Bensussan, A.; Le Bouteiller, P. Cutting edge: Soluble HLA-G1 triggers CD95/CD95 ligand-mediated apoptosis in activated CD8+ cells by interacting with CD8. J. Immunol. 2000, 164, 6100–6104. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, S.; Marquet, J.; Freeman, G.J.; Tawab, A.; Bouteiller, P.L.; Roth, P.; Bolton, W.; Ogg, G.; Boumsell, L.; Bensussan, A. Cutting edge: MHC class I triggering by a novel cell surface ligand costimulates proliferation of activated human T cells. J. Immunol. 1999, 162, 1223–1226. [Google Scholar] [PubMed]
- Frei, A.P.; Jeon, O.Y.; Kilcher, S.; Moest, H.; Henning, L.M.; Jost, C.; Pluckthun, A.; Mercer, J.; Aebersold, R.; Carreira, E.M.; et al. Direct identification of ligand-receptor interactions on living cells and tissues. Nat. Biotechnol. 2012, 30, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- Frei, A.P.; Moest, H.; Novy, K.; Wollscheid, B. Ligand-based receptor identification on living cells and tissues using TRICEPS. Nat. Protoc. 2013, 8, 1321–1336. [Google Scholar] [CrossRef]
- Hidaka, H.; Yagasaki, H.; Takahashi, Y.; Hama, A.; Nishio, N.; Tanaka, M.; Yoshida, N.; Villalobos, I.B.; Wang, Y.; Xu, Y.; et al. Increased midkine gene expression in childhood B-precursor acute lymphoblastic leukemia. Leuk. Res. 2007, 31, 1045–1051. [Google Scholar] [CrossRef]
- Robertson, M.J.; Cochran, K.J.; Cameron, C.; Le, J.M.; Tantravahi, R.; Ritz, J. Characterization of a cell line, NKL, derived from an aggressive human natural killer cell leukemia. Exp. Hematol. 1996, 24, 406–415. [Google Scholar] [PubMed]
- Celik, A.A.; Simper, G.S.; Huyton, T.; Blasczyk, R.; Bade-Doding, C. HLA-G mediated immune regulation is impaired by a single amino acid exchange in the alpha 2 domain. Hum. Immunol. 2018. [Google Scholar] [CrossRef]
- Stieglitz, F.; Celik, A.A.; von Kaisenberg, C.; Camps, M.A.; Blasczyk, R.; Bade-Doding, C. The microstructure in the placenta is influenced by the functional diversity of HLA-G allelic variants. Immunogenetics 2019, 71, 455–463. [Google Scholar] [CrossRef] [Green Version]
- Pump, W.C.; Kraemer, T.; Huyton, T.; Ho, G.T.; Blasczyk, R.; Bade-Doeding, C. Between Innate and Adaptive Immune Responses: NKG2A, NKG2C, and CD8+ T Cell Recognition of HLA-E Restricted Self-Peptides Acquired in the Absence of HLA-Ia. Int. J. Mol. Sci. 2019, 20, 1454. [Google Scholar] [CrossRef] [Green Version]
- Kaiser, B.K.; Pizarro, J.C.; Kerns, J.; Strong, R.K. Structural basis for NKG2A/CD94 recognition of HLA-E. Proc. Natl. Acad. Sci. USA 2008, 105, 6696–6701. [Google Scholar] [CrossRef] [Green Version]
- Hickman-Miller, H.D.; Hildebrand, W.H. The immune response under stress: The role of HSP-derived peptides. Trends Immunol. 2004, 25, 427–433. [Google Scholar] [CrossRef]
- Velardi, A.; Ruggeri, L.; Mancusi, A.; Aversa, F.; Christiansen, F.T. Natural killer cell allorecognition of missing self in allogeneic hematopoietic transplantation: A tool for immunotherapy of leukemia. Curr. Opin. Immunol. 2009, 21, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Minetto, P.; Guolo, F.; Pesce, S.; Greppi, M.; Obino, V.; Ferretti, E.; Sivori, S.; Genova, C.; Lemoli, R.M.; Marcenaro, E. Harnessing NK Cells for Cancer Treatment. Front. Immunol. 2019, 10, 2836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, J.S.; Soignier, Y.; Panoskaltsis-Mortari, A.; McNearney, S.A.; Yun, G.H.; Fautsch, S.K.; McKenna, D.; Le, C.; Defor, T.E.; Burns, L.J.; et al. Successful adoptive transfer and in vivo expansion of human haploidentical NK cells in patients with cancer. Blood 2005, 105, 3051–3057. [Google Scholar] [CrossRef] [Green Version]
- Tonn, T.; Schwabe, D.; Klingemann, H.G.; Becker, S.; Esser, R.; Koehl, U.; Suttorp, M.; Seifried, E.; Ottmann, O.G.; Bug, G. Treatment of patients with advanced cancer with the natural killer cell line NK-92. Cytotherapy 2001, 15, 1563–1570. [Google Scholar] [CrossRef] [PubMed]
- Arai, S.; Meagher, R.; Swearingen, M.; Myint, H.; Rich, E.; Martinson, J.; Klingemann, H. Infusion of the allogeneic cell line NK-92 in patients with advanced renal cell cancer or melanoma: A phase I trial. Cytotherapy 2008, 10, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Vacca, P.; Pietra, G.; Tumino, N.; Munari, E.; Mingari, M.C.; Moretta, L. Exploiting Human NK Cells in Tumor Therapy. Front. Immunol. 2020, 10, 3013. [Google Scholar] [CrossRef]
- Holsken, O.; Miller, M.; Cerwenka, A. Exploiting natural killer cells for therapy of melanoma. J. Dtsch. Dermatol. Ges. 2014, 13, 23–29. [Google Scholar] [CrossRef]
- Gillard-Bocquet, M.; Caer, C.; Cagnard, N.; Crozet, L.; Perez, M.; Fridman, W.H.; Sautes-Fridman, C.; Cremer, I. Lung tumor microenvironment induces specific gene expression signature in intratumoral NK cells. Front. Immunol. 2013, 4, 19. [Google Scholar] [CrossRef] [Green Version]
- Platonova, S.; Cherfils-Vicini, J.; Damotte, D.; Crozet, L.; Vieillard, V.; Validire, P.; Andre, P.; Dieu-Nosjean, M.C.; Alifano, M.; Regnard, J.F.; et al. Profound coordinated alterations of intratumoral NK cell phenotype and function in lung carcinoma. Cancer Res. 2011, 71, 5412–5422. [Google Scholar] [CrossRef] [Green Version]
- Kamiya, T.; Seow, S.V.; Wong, D.; Robinson, M.; Campana, D. Blocking expression of inhibitory receptor NKG2A overcomes tumor resistance to NK cells. J. Clin. Investig. 2019, 129, 2094–2106. [Google Scholar] [CrossRef] [Green Version]
- Braud, V.M.; Allan, D.S.; O’Callaghan, C.A.; Soderstrom, K.; D’Andrea, A.; Ogg, G.S.; Lazetic, S.; Young, N.T.; Bell, J.I.; Phillips, J.H.; et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 1998, 391, 795–799. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Wang, Z.; Chen, X.; Tao, A.; He, L.; Fu, S.; Zhang, Z.; Fu, Y.; Guo, C.; Liu, J.; et al. NKG2C+NKG2A− Natural Killer Cells are Associated with a Lower Viral Set Point and may Predict Disease Progression in Individuals with Primary HIV Infection. Front. Immunol. 2017, 8, 1176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beziat, V.; Hervier, B.; Achour, A.; Boutolleau, D.; Marfain-Koka, A.; Vieillard, V. Human NKG2A overrides NKG2C effector functions to prevent autoreactivity of NK cells. Blood 2011, 117, 4394–4396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saez-Borderias, A.; Romo, N.; Magri, G.; Guma, M.; Angulo, A.; Lopez-Botet, M. IL-12-dependent inducible expression of the CD94/NKG2A inhibitory receptor regulates CD94/NKG2C+ NK cell function. J. Immunol. 2009, 182, 829–836. [Google Scholar] [CrossRef] [Green Version]
- Kusumi, M.; Yamashita, T.; Fujii, T.; Nagamatsu, T.; Kozuma, S.; Taketani, Y. Expression patterns of lectin-like natural killer receptors, inhibitory CD94/NKG2A, and activating CD94/NKG2C on decidual CD56bright natural killer cells differ from those on peripheral CD56dim natural killer cells. J. Reprod. Immunol. 2006, 70, 33–42. [Google Scholar] [CrossRef]
- Bade-Doeding, C.; Cano, P.; Huyton, T.; Badrinath, S.; Eiz-Vesper, B.; Hiller, O.; Blasczyk, R. Mismatches outside exons 2 and 3 do not alter the peptide motif of the allele group B*44:02P. Hum. Immunol. 2011, 72, 1039–1044. [Google Scholar] [CrossRef]
- Kunze-Schumacher, H.; Blasczyk, R.; Bade-Doeding, C. Soluble HLA technology as a strategy to evaluate the impact of HLA mismatches. J. Immunol. Res. 2014, 2014, 246171. [Google Scholar] [CrossRef]
| Cells | HLA-G | Method | |
|---|---|---|---|
| ILT2 | NK cells, T cells, DCs, and decidual macrophages | sHLA-G1 | Crystal structure [52] |
| sHLA-G1 | Crystal structure [53] | ||
| sHLA-G1 | Surface plasmon resonance [35] | ||
| sHLA-G1 | Surface plasmon resonance [54] | ||
| HLA-G tetramers | Tetramer-binding assays [55] | ||
| mHLA-G1 | Cytotoxicity assays [33] | ||
| ILT4 | monocytes, macrophages, and DCs | β2m-free HLA-G1 dimers | Surface plasmon resonance [52] |
| sHLA-G1 | Crystal structure [56] | ||
| sHLA-G1 | Surface plasmon resonance [35] | ||
| HLA-G tetramers | Tetramer-binding assays [55] | ||
| sHLA-G | Cell binding assays [57] | ||
| KIR2DL4 | NK cells | mHLA-G | Binding assays and cytotoxicity assays [36] |
| mHLA-G | Cytotoxicity assays [58] | ||
| mHLA-G | Binding assays [59] | ||
| CD8 | CD8+ T cells | sHLA-G1 | Apoptosis assay [60] |
| CD160 | Endothelial cells | sHLA-G1 | Radiolabeled cell-binding competition assay, tetramer-binding, antibody-blocking [39] |
| mHLA-G | Conjugate formation (cell binding assay) of CHO-CD160 transfectants and HLA-expressing cells [61] |
| Protein | Log2FC | −Log10(adj. p-Value) | |
|---|---|---|---|
| MOT2 | Monocarboxylate transporter 2 | −2.95164 | 2.948745312 |
| S43A3 | Solute carrier family 43 member 3 | −1.19412 | 3.325027107 |
| MOT1 | Monocarboxylate transporter 1 | −1.8302 | 5.028116407 |
| PPIB | Peptidyl-prolyl cis-trans isomerase | 1.274108 | 7.824816967 |
| MK | Midkine | 5.897202 | 10.51019622 |
| B2MG | β2m | 2.770848 | 11.09377281 |
| TFR1 | Transferrin receptor protein 1 | −1.64478 | 12.72159366 |
| MOT4 | Monocarboxylate transporter 4 | −2.89668 | 12.72159366 |
| HLAG | HLA-G | 4.550522 | 12.72159366 |
| TRFE | Transferrin | −6.51903 | 12.72159366 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hò, G.-G.T.; Celik, A.A.; Huyton, T.; Hiemisch, W.; Blasczyk, R.; Simper, G.S.; Bade-Doeding, C. NKG2A/CD94 Is a New Immune Receptor for HLA-G and Distinguishes Amino Acid Differences in the HLA-G Heavy Chain. Int. J. Mol. Sci. 2020, 21, 4362. https://doi.org/10.3390/ijms21124362
Hò G-GT, Celik AA, Huyton T, Hiemisch W, Blasczyk R, Simper GS, Bade-Doeding C. NKG2A/CD94 Is a New Immune Receptor for HLA-G and Distinguishes Amino Acid Differences in the HLA-G Heavy Chain. International Journal of Molecular Sciences. 2020; 21(12):4362. https://doi.org/10.3390/ijms21124362
Chicago/Turabian StyleHò, Gia-Gia T., Alexander A. Celik, Trevor Huyton, Wiebke Hiemisch, Rainer Blasczyk, Gwendolin S. Simper, and Christina Bade-Doeding. 2020. "NKG2A/CD94 Is a New Immune Receptor for HLA-G and Distinguishes Amino Acid Differences in the HLA-G Heavy Chain" International Journal of Molecular Sciences 21, no. 12: 4362. https://doi.org/10.3390/ijms21124362






