A Comparison of the Flavonoid Biosynthesis Mechanisms of Dendrobium Species by Analyzing the Transcriptome and Metabolome
Abstract
:1. Introduction
2. Results
2.1. Determination of Flavonoids and Alkaloids Contents
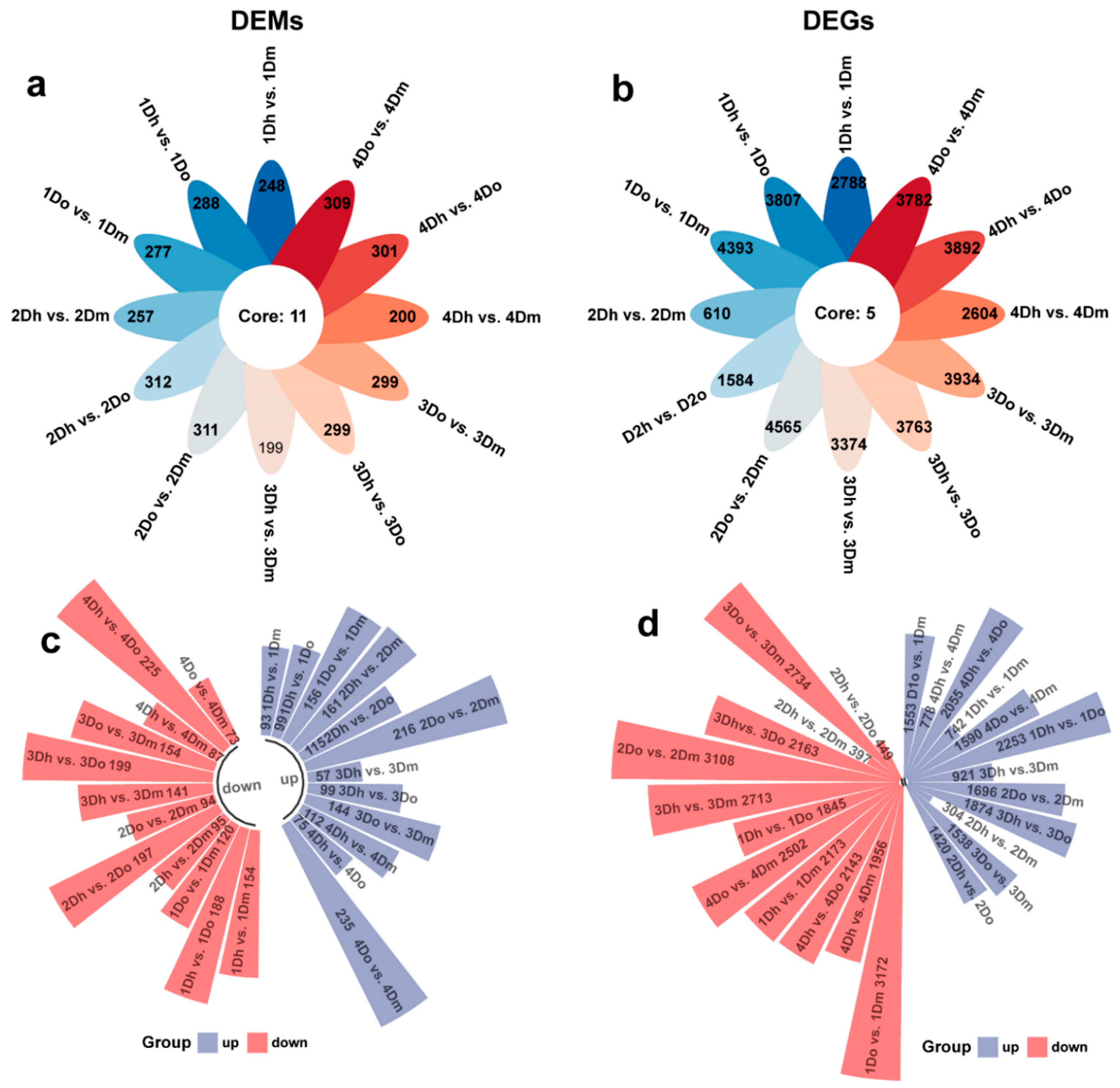
2.2. Identification of DAMs and DEGs
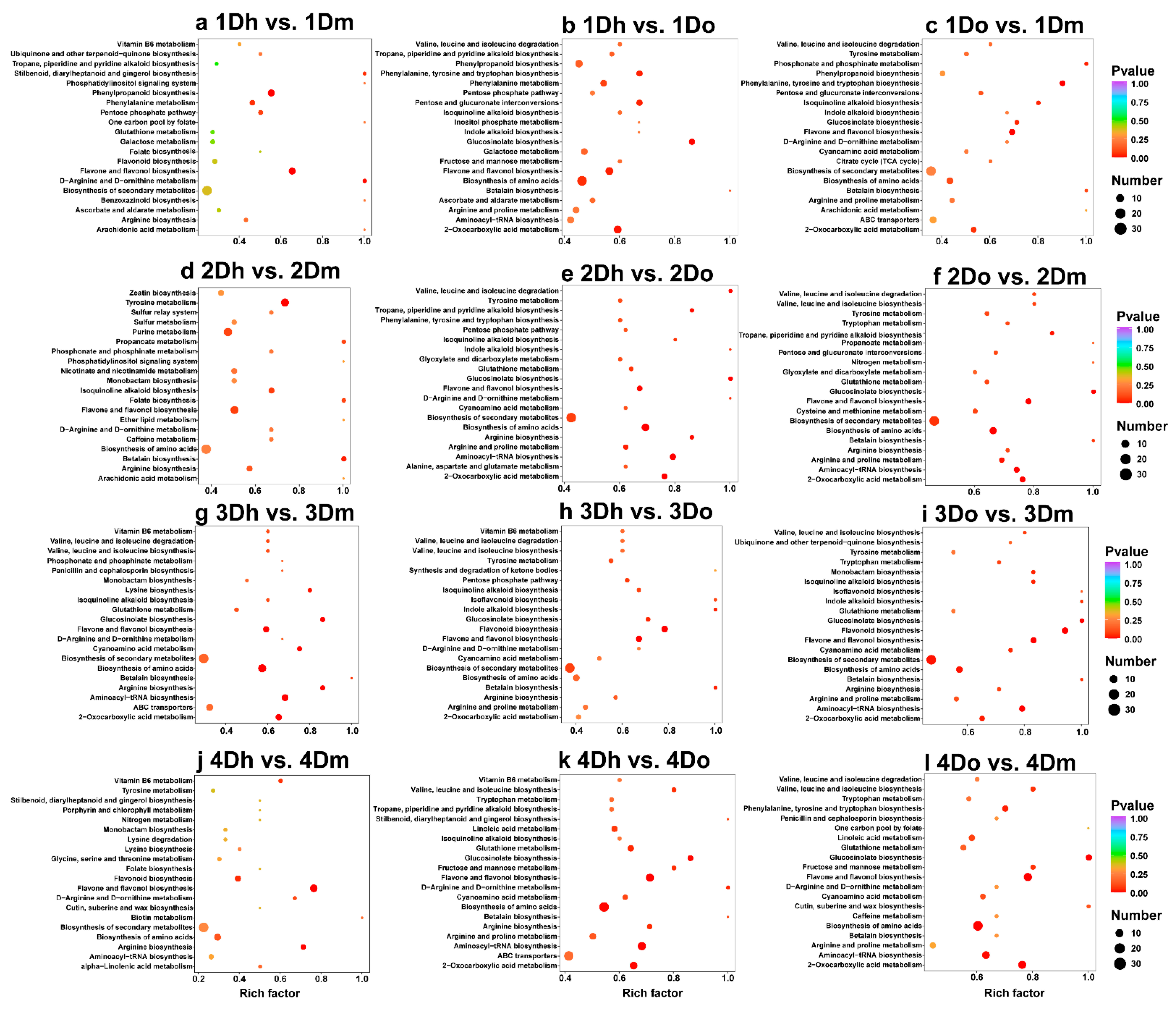
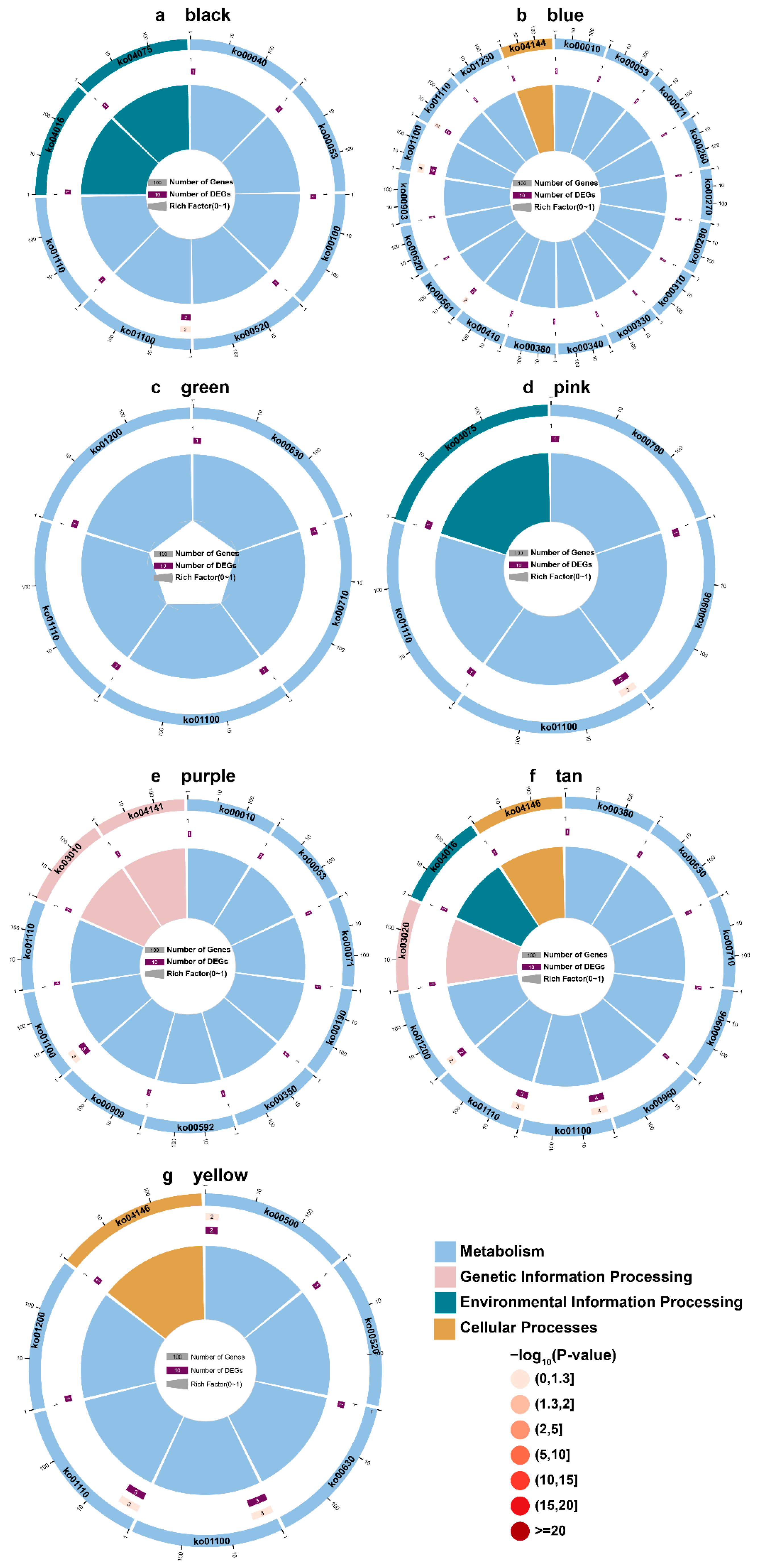
2.3. KEGG Enrichment of DAMs
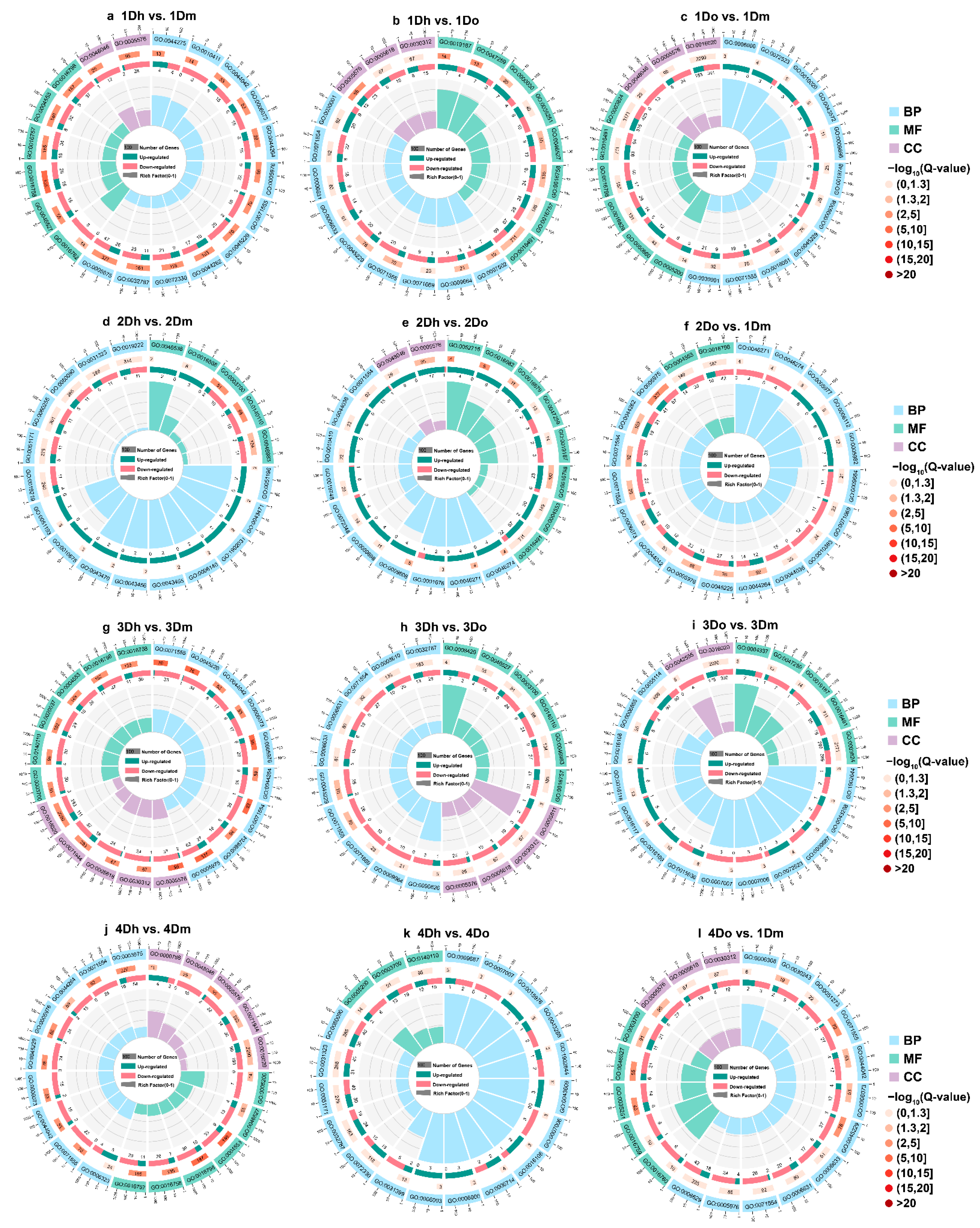
2.4. GO Enrichment of DEGs
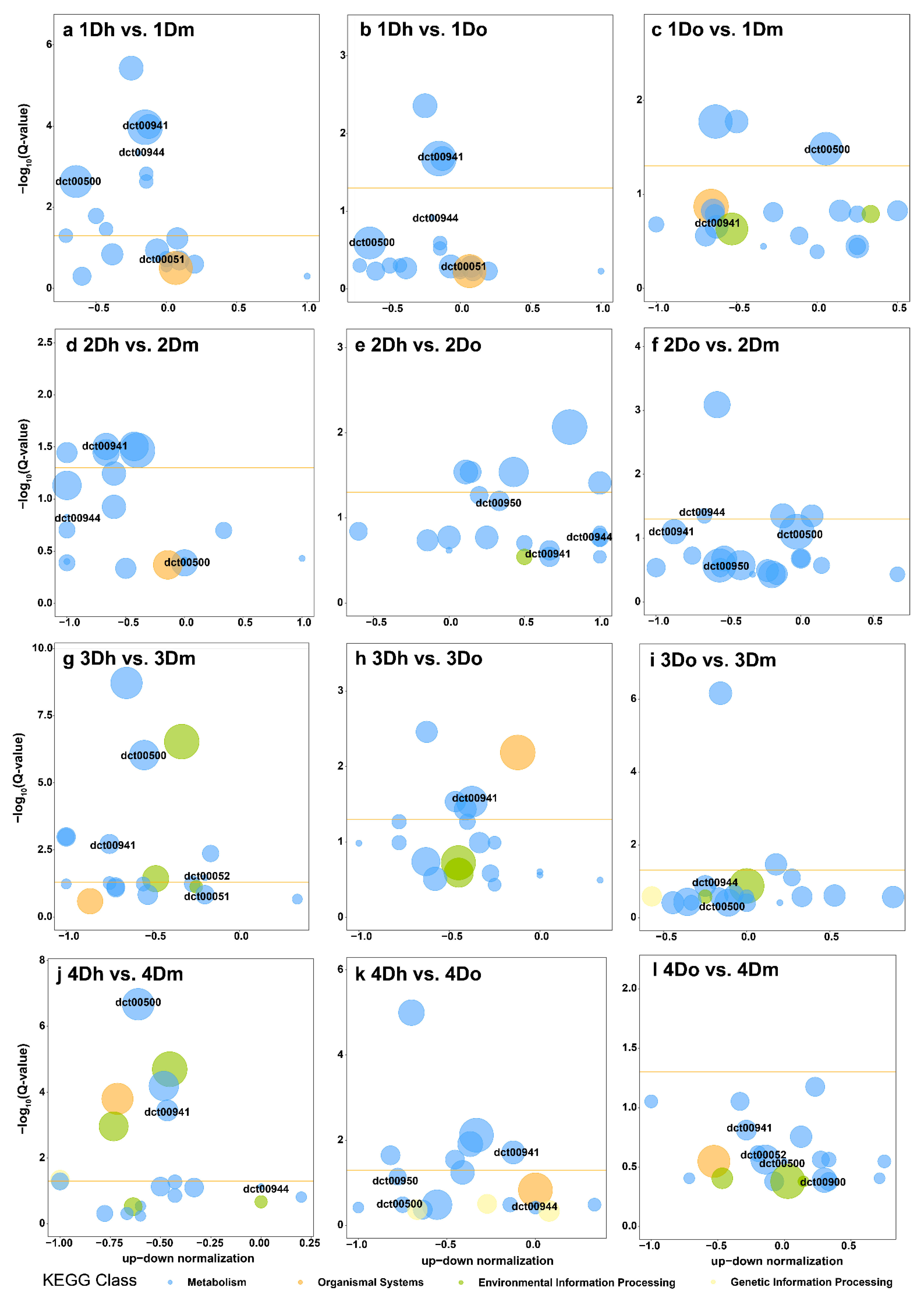
2.5. KEGG Enrichment of DEGs
2.6. Integrative Analysis of DAMs and DEGs
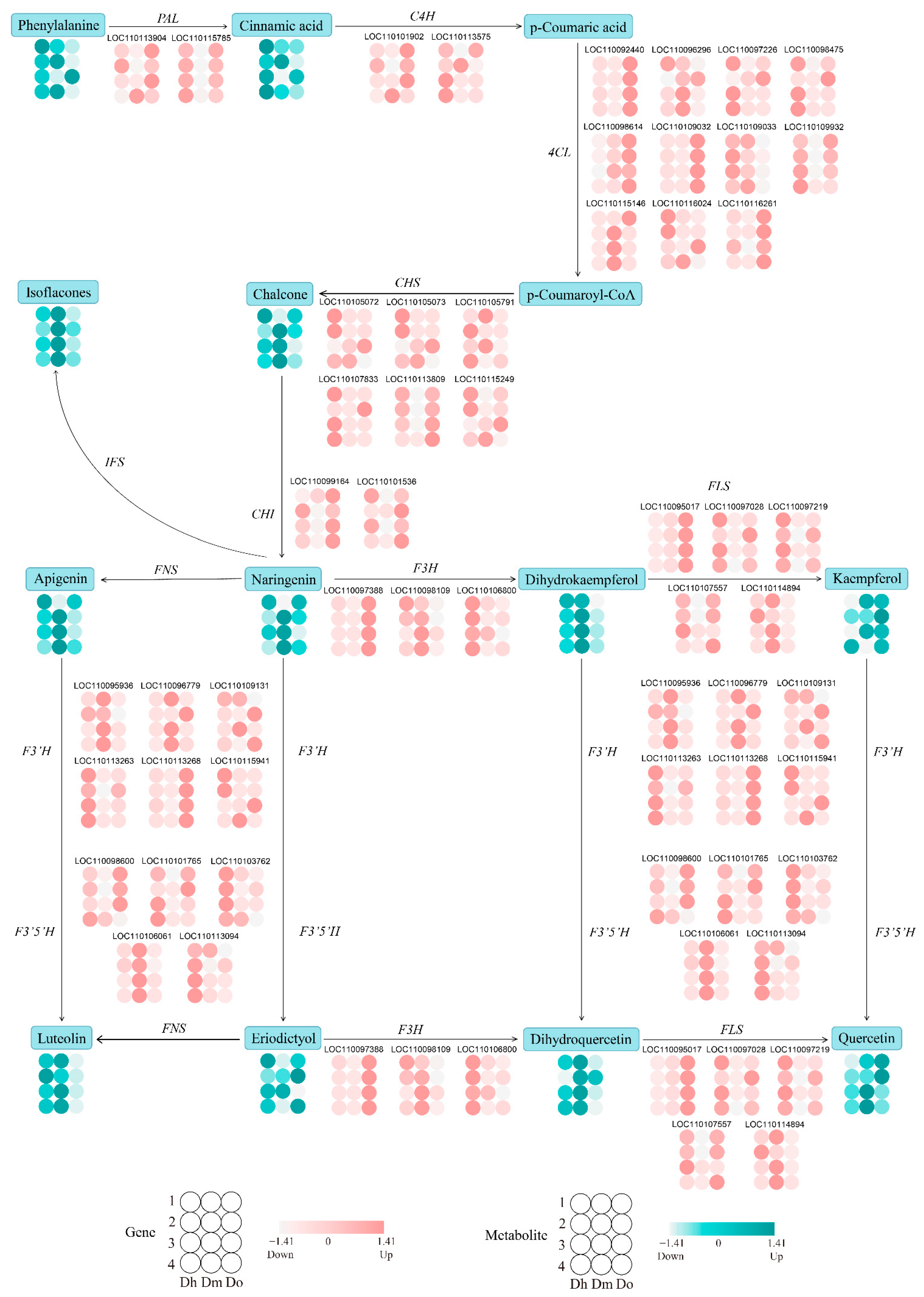
2.7. Changes of Genes and Metabolites in Regulatory Networks for Flavonoid Biosynthesis
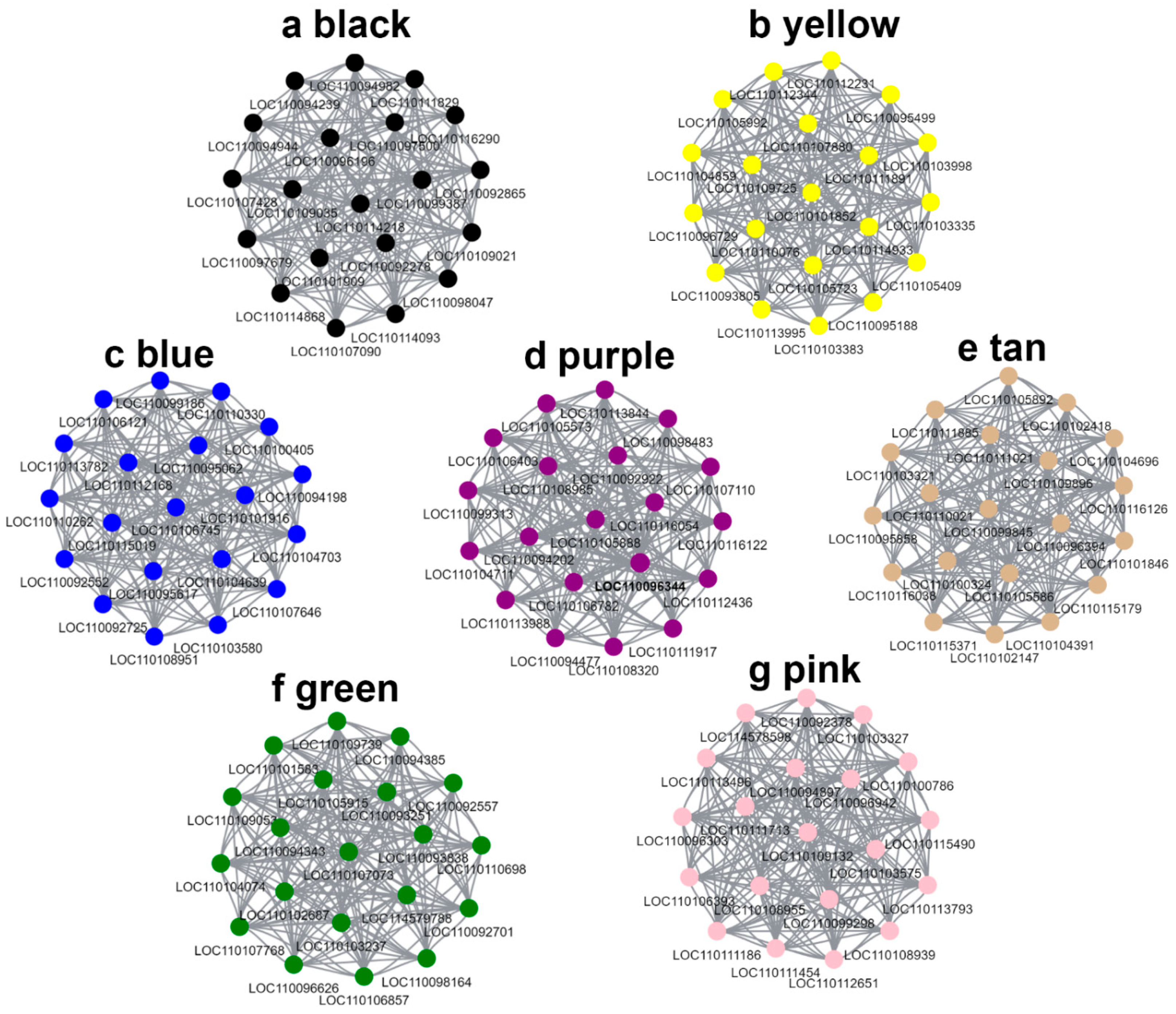
2.8. Weighted Gene Co-Expression Network Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Sample Collection
4.2. Determination of Total Flavonoid and Total Alkaloid Contents
4.3. Widely Targeted Metabolomics Profiling
4.4. RNA Extraction, Illumina Sequencing, and Analysis of DEGs
4.5. Gene Co-Expression Network Construction
4.6. Combination Analysis of Transcriptome and Metabolome
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, Z.; Zhao, M.; Cui, H.; Li, J.; Wang, M. Transcriptomic Landscape of Medicinal Dendrobium Reveals Genes Associated With the Biosynthesis of Bioactive Components. Front. Plant Sci. 2020, 11, 391. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Yao, Y.; Cai, Y.; Lin, Y. Molecular cloning and sequence analysis of a phenylalanine ammonia-lyase gene from dendrobium. PLoS ONE 2013, 8, e62352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, S.; He, R.; Yang, S.; Chen, Z.; Jiang, M.; Lu, J.; Wang, H. Start codon targeted (SCoT) and target region amplification polymorphism (TRAP) for evaluating the genetic relationship of Dendrobium species. Gene 2015, 567, 182–188. [Google Scholar] [CrossRef]
- Zhang, G.-Q.; Xu, Q.; Bian, C.; Tsai, W.-C.; Yeh, C.-M.; Liu, K.-W.; Yoshida, K.; Zhang, L.-S.; Chang, S.-B.; Chen, F.; et al. The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 2016, 6, 19029. [Google Scholar] [CrossRef] [Green Version]
- Zha, X.-Q.; Pan, L.-H.; Luo, J.-P.; Wang, J.-H.; Wei, P.; Bansal, V. Enzymatic fingerprints of polysaccharides of Dendrobium officinale and their application in identification of Dendrobium species. J. Nat. Med. 2012, 66, 525–534. [Google Scholar] [CrossRef]
- Jin, Q.; Jiao, C.Y.; Sun, S.W.; Song, C.; Cai, Y.P.; Lin, Y.; Fan, H.H.; Zhu, Y.F. Metabolic Analysis of Medicinal Dendrobium officinale and Dendrobium huoshanense during Different Growth Years. PLoS ONE 2016, 11, e0146607. [Google Scholar] [CrossRef]
- Yuan, Y.; Yu, M.; Jia, Z.; Song, X.; Liang, Y.; Zhang, J. Analysis of Dendrobium huoshanense transcriptome unveils putative genes associated with active ingredients synthesis. BMC Genom. 2018, 19, 978. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhang, J.; Kallman, J.; Liu, X.; Meng, M.; Lin, J. Polysaccharide biosynthetic pathway profiling and putative gene mining of Dendrobium moniliforme using RNA-Seq in different tissues. BMC Plant Biol. 2019, 19, 521. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.; Zhang, B.; Tang, X.; Zhang, J.; Lin, J. Comparative Transcriptome Analysis of Different Dendrobium Species Reveals Active Ingredients-Related Genes and Pathways. Int. J. Mol. Sci. 2020, 21, 861. [Google Scholar] [CrossRef] [Green Version]
- Baek, J.M.; Kim, J.-Y.; Ahn, S.-J.; Cheon, Y.-H.; Yang, M.; Oh, J.; Choi, M.K. Dendrobium moniliforme Exerts Inhibitory Effects on Both Receptor Activator of Nuclear Factor Kappa-B Ligand-Mediated Osteoclast Differentiation in Vitro and Lipopolysaccharide-Induced Bone Erosion in Vivo. Molecules 2016, 21, 295. [Google Scholar] [CrossRef]
- Paudel, M.R.; Chand, M.B.; Pant, B.; Pant, B. Antioxidant and cytotoxic activities of Dendrobium moniliforme extracts and the detection of related compounds by GC-MS. BMC Complement. Altern. Med. 2018, 18, 134. [Google Scholar] [CrossRef] [Green Version]
- Yue, H.; Zeng, H.; Ding, K. A review of isolation methods, structure features and bioactivities of polysaccharides from Dendrobium species. Chin. J. Nat. Med. 2020, 18, 1–27. [Google Scholar] [CrossRef]
- Zheng, S.-G.; Hu, Y.-D.; Zhao, R.-X.; Yan, S.; Zhang, X.-Q.; Zhao, T.-M.; Chun, Z. Genome-wide researches and applications on Dendrobium. Planta 2018, 248, 769–784. [Google Scholar] [CrossRef] [PubMed]
- Paudel, M.R.; Chand, M.B.; Pant, B.; Pant, B. Assessment of Antioxidant and Cytotoxic Activities of Extracts of Dendrobium crepidatum. Biomolecules 2019, 9, 478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mou, Z.; Zhao, Y.; Ye, F.; Shi, Y.; Kennelly, E.J.; Chen, S.; Zhao, D. Identification, Biological Activities and Biosynthetic Pathway of Dendrobium Alkaloids. Front. Pharmacol. 2021, 12, 605994. [Google Scholar] [CrossRef]
- Zuo, J.; Zu, M.; Liu, L.; Song, X.; Yuan, Y. Composition and diversity of bacterial communities in the rhizosphere of the Chinese medicinal herb Dendrobium. BMC Plant Biol. 2021, 21, 127. [Google Scholar] [CrossRef]
- He, C.; Wu, K.; Zhang, J.; Liu, X.; Zeng, S.; Yu, Z.; Zhang, X.; Teixeira da Silva, J.A.; Deng, R.; Tan, J.; et al. Cytochemical Localization of Polysaccharides in Dendrobium officinale and the Involvement of DoCSLA6 in the Synthesis of Mannan Polysaccharides. Front. Plant Sci. 2017, 8, 173. [Google Scholar] [CrossRef] [Green Version]
- He, C.; Yu, Z.; Teixeira da Silva, J.A.; Zhang, J.; Liu, X.; Wang, X.; Zhang, X.; Zeng, S.; Wu, K.; Tan, J.; et al. DoGMP1 from Dendrobium officinale contributes to mannose content of water-soluble polysaccharides and plays a role in salt stress response. Sci. Rep. 2017, 7, 41010. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Li, Y.; Li, C.; Luo, H.; Wang, L.; Qian, J.; Luo, X.; Xiang, L.; Song, J.; Sun, C.; et al. Analysis of the Dendrobium officinale transcriptome reveals putative alkaloid biosynthetic genes and genetic markers. Gene 2013, 527, 131–138. [Google Scholar] [CrossRef]
- Liu, H.-J.; Jiang, X.-X.; Guo, Y.-Z.; Sun, F.-H.; Kou, X.-H.; Bao, Y.; Zhang, Z.-Q.; Lin, Z.-H.; Ding, T.-B.; Jiang, L.; et al. The flavonoid TL-2-8 induces cell death and immature mitophagy in breast cancer cells via abrogating the function of the AHA1/Hsp90 complex. Acta Pharmacol. Sin. 2017, 38, 1381–1393. [Google Scholar] [CrossRef]
- Olayinka, E.T.; Ore, A.; Adeyemo, O.A.; Ola, O.S. The role of flavonoid antioxidant, morin in improving procarbazine-induced oxidative stress on testicular function in rat. Porto Biomed. J. 2019, 4, e28. [Google Scholar] [CrossRef] [PubMed]
- Dodd, G.F.; Williams, C.M.; Butler, L.T.; Spencer, J.P.E. Acute effects of flavonoid-rich blueberry on cognitive and vascular function in healthy older adults. Nutr. Healthy Aging 2019, 5, 119–132. [Google Scholar] [CrossRef] [Green Version]
- Frandsen, J.R.; Narayanasamy, P. Neuroprotection through flavonoid: Enhancement of the glyoxalase pathway. Redox Biol. 2018, 14, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Alqurashi, R.M.; Galante, L.A.; Rowland, I.R.; Spencer, J.P.; Commane, D.M. Consumption of a flavonoid-rich açai meal is associated with acute improvements in vascular function and a reduction in total oxidative status in healthy overweight men. Am. J. Clin. Nutr. 2016, 104, 1227–1235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, X.; Shang, E.; Zhou, G.; Tang, Y.; Guo, S.; Su, S.; Jin, C.; Qian, D.; Qin, Y.; Duan, J.-A. Comparative characterization of total flavonol glycosides and terpene lactones at different ages, from different cultivation sources and genders of Ginkgo biloba leaves. Int. J. Mol. Sci. 2012, 13, 10305–10315. [Google Scholar] [CrossRef]
- Wang, Q.; Jiang, Y.; Mao, X.; Yu, W.; Lu, J.; Wang, L. Integration of morphological, physiological, cytological, metabolome and transcriptome analyses reveal age inhibited accumulation of flavonoid biosynthesis in Ginkgo biloba leaves. Ind. Crops Prod. 2022, 187, 115405. [Google Scholar] [CrossRef]
- Naghiloo, S.; Movafeghi, A.; Delazar, A.; Nazemiyeh, H.; Asnaashari, S.; Dadpour, M.R. Ontogenetic variation of volatiles and antioxidant activity in leaves of Astragalus compactus Lam. (Fabaceae). EXCLI J. 2012, 11, 436–443. [Google Scholar]
- Shi, W.; Wang, Y.; Li, J.; Zhang, H.; Ding, L. Investigation of ginsenosides in different parts and ages of Panax ginseng. Food Chem. 2007, 102, 664–668. [Google Scholar] [CrossRef]
- Zhu, Y.; Xu, C.-H.; Huang, J.; Li, G.-Y.; Liu, X.-H.; Sun, S.-Q.; Wang, J.-H. Rapid discrimination of cultivated Codonopsis lanceolata in different ages by FT-IR and 2DCOS-IR. J. Mol. Struct. 2014, 1069, 272–279. [Google Scholar] [CrossRef]
- Yuan, Y.; Yu, M.; Zhang, B.; Liu, X.; Zhang, J. Comparative nutritional characteristics of the three major Chinese Dendrobium species with different growth years. PLoS ONE 2019, 14, e0222666. [Google Scholar] [CrossRef]
- Chen, N.-D.; Chen, N.-F.; Li, J.; Cao, C.-Y.; Wang, J.-M.; Huang, H.-P. Similarity Evaluation of Different Origins and Species of Dendrobiums by GC-MS and FTIR Analysis of Polysaccharides. Int. J. Anal. Chem. 2015, 2015, 713410. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Chen, F.; Cheng, H.; Huang, G. Modification and application of polysaccharide from traditional Chinese medicine such as Dendrobium officinale. Int. J. Biol. Macromol. 2020, 157, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Jiang, W.; Liu, Y.; Meng, X.; Su, X.; Cao, M.; Wu, L.; Yu, N.; Xing, S.; Peng, D. Putative genes in alkaloid biosynthesis identified in Dendrobium officinale by correlating the contents of major bioactive metabolites with genes expression between Protocorm-like bodies and leaves. BMC Genom. 2021, 22, 579. [Google Scholar] [CrossRef] [PubMed]
- Belkheir, A.K.; Gaid, M.; Liu, B.Y.; Hansch, R.; Beerhues, L. Benzophenone Synthase and Chalcone Synthase Accumulate in the Mesophyll of Hypericum perforatum Leaves at Different Developmental Stages. Front. Plant Sci. 2016, 7, 921. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Yu, Y.; Shi, R.; Xie, G.; Zhu, Y.; Wu, G.; Qin, M. Organ-Specific Metabolic Shifts of Flavonoids in Scutellaria baicalensis at Different Growth and Development Stages. Molecules 2018, 23, 428. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Liu, M.; Shi, K.; Yu, Z.; Zhou, Y.; Fan, R.; Shi, Q. Dynamic Changes in Metabolite Accumulation and the Transcriptome during Leaf Growth and Development in Eucommia ulmoides. Int. J. Mol. Sci. 2019, 20, 4030. [Google Scholar] [CrossRef] [Green Version]
- Kong, D.-X.; Li, Y.-Q.; Bai, M.; He, H.-J.; Liang, G.-X.; Wu, H. Correlation between the dynamic accumulation of the main effective components and their associated regulatory enzyme activities at different growth stages in Lonicera japonica Thunb. Ind. Crop. Prod. 2017, 96, 16–22. [Google Scholar] [CrossRef]
- Hu, G.Q.; Yuan, Y.; Wu, C.; Jiang, C.; Wang, Z.Y.; Lin, S.F.; Wu, Z.G. Effects of different development stages on growth and accumulation of active components of scutellaria baicalensis. China J. Chin. Mater. Med. 2012, 37, 3793–3798. [Google Scholar]
- Jang, G.H.; Kim, H.W.; Lee, M.K.; Jeong, S.Y.; Bak, A.R.; Lee, D.J.; Kim, J.B. Characterization and quantification of flavonoid glycosides in the Prunus genus by UPLC-DAD-QTOF/MS. Saudi J. Biol. Sci. 2018, 25, 1622–1631. [Google Scholar] [CrossRef]
- Zhou, C.; Mei, X.; Rothenberg, D.O.; Yang, Z.; Zhang, W.; Wan, S.; Yang, H.; Zhang, L. Metabolome and Transcriptome Analysis Reveals Putative Genes Involved in Anthocyanin Accumulation and Coloration in White and Pink Tea (Camellia sinensis) Flower. Molecules 2020, 25, 190. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Jiang, X.; Chen, L.; Li, W.-W.; Lai, S.; Fu, Z.; Liu, Y.; Qian, Y.; Gao, L.; Xia, T. Functional Analyses of Flavonol Synthase Genes From Camellia sinensis Reveal Their Roles in Anther Development. Front. Plant Sci. 2021, 12, 753131. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Huang, S.; Zhou, Y.; Wang, X.; Liu, A.; Hou, B.; Ye, M. Analysis of the genetic relationships among 3 Dendrobium species based on genotyping by sequencing. Chin. Wild Plant Resour. 2021, 40, 19–24. [Google Scholar]
- Guo, L.; Gao, L.; Ma, X.; Guo, F.; Ruan, H.; Bao, Y.; Xia, T.; Wang, Y. Functional analysis of flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylases from tea plant (Camellia sinensis), involved in the B-ring hydroxylation of flavonoids. Gene 2019, 717, 144046. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Huang, H.; Meng, L.; Hu, K.; Dai, S.-L. Isolation and functional analysis of a homolog of flavonoid 3′,5′-hydroxylase gene from Pericallis × hybrida. Physiol. Plant. 2013, 149, 151–159. [Google Scholar] [CrossRef]
- Nguyen, Y.T.H.; Hoang, H.T.T.; Mai, A.T.H.; Nguyen, L.T.N.; Nguyen, Q.H.; Pham, N.T.T.; Sy, T.D.; Chu, M.H. The Aconitum carmichaelii F3′5′H Gene Overexpression Increases Flavonoid Accumulation in Transgenic Tobacco Plants. Horticulturae 2021, 7, 384. [Google Scholar] [CrossRef]
- Sahagun, J.; Ratanasut, K. Development of Flavanone-3-hydroxylase (F3′H) Gene Silencing System in Dendrobium Sonia Earsakul Flowers for Engineering Aurone Biosynthetic Pathway. In Proceedings of the International Biochemistry & Molecular Biology Conference, Songkla, Thailand, 26–27 May 2016; pp. 266–269. [Google Scholar]
- Kriangphan, N.; Vuttipongchaikij, S.; Kittiwongwattana, C.; Suttangkakul, A.; Pinmanee, P.; Sakulsathaporn, A.; Suwimon, R.; Suputtitada, S.; Chanvivattana, Y.; Apisitwanich, S. Effects of sequence and expression of eight anthocyanin biosynthesis genes on floral coloration in four Dendrobium hybrids. Horticutural J. 2015, 84, 83–92. [Google Scholar] [CrossRef] [Green Version]
- Whang, S.S.; Um, W.S.; Song, I.-J.; Lim, P.O.; Choi, K.; Park, K.-W.; Kang, K.-W.; Choi, M.S.; Koo, J.C. Molecular analysis of anthocyanin biosynthetic genes and control of flower coloration by flavonoid 3′, 5′-hydroxylase (F3′ 5′ H) in Dendrobium moniliforme. J. Plant Biol. 2011, 54, 209–218. [Google Scholar] [CrossRef]
- Bai, C.; Yang, J.; Cao, B.; Xue, Y.; Gao, P.; Liang, H.; Li, G. Growth years and post-harvest processing methods have critical roles on the contents of medicinal active ingredients of Scutellaria baicalensis. Ind. Crops Prod. 2020, 158, 112985. [Google Scholar] [CrossRef]
- Tapas, A.R.; Sakarkar, D.; Kakde, R. Flavonoids as nutraceuticals: A review. Trop. J. Pharm. Res. 2008, 7, 1089–1099. [Google Scholar] [CrossRef] [Green Version]
- Ren, J.; Lu, Y.; Qian, Y.; Chen, B.; Wu, T.; Ji, G. Recent progress regarding kaempferol for the treatment of various diseases. Exp. Ther. Med. 2019, 18, 2759–2776. [Google Scholar] [CrossRef] [Green Version]
- Imran, M.; Salehi, B.; Sharifi-Rad, J.; Aslam Gondal, T.; Saeed, F.; Imran, A.; Shahbaz, M.; Tsouh Fokou, P.V.; Umair Arshad, M.; Khan, H.; et al. Kaempferol: A Key Emphasis to Its Anticancer Potential. Molecules 2019, 24, 2277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, S.-M.; Deng, X.-T.; Zhou, J.; Li, Q.-P.; Ge, X.-X.; Miao, L. Pharmacological basis and new insights of quercetin action in respect to its anti-cancer effects. Biomed Pharmacother. 2020, 121, 109604. [Google Scholar] [CrossRef] [PubMed]
- Vafadar, A.; Shabaninejad, Z.; Movahedpour, A.; Fallahi, F.; Taghavipour, M.; Ghasemi, Y.; Akbari, M.; Shafiee, A.; Hajighadimi, S.; Moradizarmehri, S.; et al. Quercetin and cancer: New insights into its therapeutic effects on ovarian cancer cells. Cell Biosci. 2020, 10, 32. [Google Scholar] [CrossRef]
- Deng, Z.; Hassan, S.; Rafiq, M.; Li, H.; He, Y.; Cai, Y.; Kang, X.; Liu, Z.; Yan, T. Pharmacological Activity of Eriodictyol: The Major Natural Polyphenolic Flavanone. Evid.-Based Complement. Altern. Med. 2020, 2020, 6681352. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.A. The leaf flavonoids of the Orchidaceae. Phytochemistry 1979, 18, 803–813. [Google Scholar] [CrossRef]
- ZENG, Y.-Y.; NIE, X.-T.; LI, Z.-J.; ZHANG, M.; YANG, Y.-B.; WANG, W.; SUN, Z.-Y. Research progress on active ingredients of flavonoids in traditional Chinese medicine Dendrobium. Chin. J. Exp. Tradit. Med. Formulae 2021, 24, 197–206. [Google Scholar]
- Zhou, C.; Luo, Y.; Lei, Z.; Wei, G. UHPLC-ESI-MS Analysis of purified flavonoids fraction from stem of Dendrobium denneaum Paxt. and its preliminary study in inducing apoptosis of HepG2 Cells. Evid.-Based Complement. Altern. Med. 2018, 2018, 8936307. [Google Scholar] [CrossRef] [Green Version]
- Bouyahya, A.; Abrini, J.; El-Baabou, A.; Dakka, Y.B.A.N. Determination of Phenol Content and Antibacterial Activity of Five Medicinal Plants Ethanolic Extracts from North-West of Morocco. J. Plant Pathol. Microbiol. 2016, 7, 4. [Google Scholar] [CrossRef]
- Yuan, Y.; Zuo, J.; Zhang, H.; Zu, M.; Yu, M.; Liu, S. Transcriptome and metabolome profiling unveil the accumulation of flavonoids in Dendrobium officinale. Genomics 2022, 114, 110324. [Google Scholar] [CrossRef]
- Yuan, Y.; Zuo, J.; Zhang, H.; Li, R.; Yu, M.; Liu, S. Integration of Transcriptome and Metabolome Provides New Insights to Flavonoids Biosynthesis in Dendrobium huoshanense. Front. Plant Sci. 2022, 13, 850090. [Google Scholar] [CrossRef]
- Yuan, Y.; Zuo, J.; Zhang, H.; Zu, M.; Liu, S. Analysis of the different growth years accumulation of flavonoids in Dendrobium moniliforme (L.) Sw. by the integration of metabolomic and transcriptomic approaches. Front. Nutr. 2022, 9, 2284. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Saito, R.; Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.-L.; Lotia, S.; Pico, A.R.; Bader, G.D.; Ideker, T. A travel guide to Cytoscape plugins. Nat. Methods 2012, 9, 1069–1076. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Zhang, H.; Yuan, Y. A Comparison of the Flavonoid Biosynthesis Mechanisms of Dendrobium Species by Analyzing the Transcriptome and Metabolome. Int. J. Mol. Sci. 2022, 23, 11980. https://doi.org/10.3390/ijms231911980
Liu S, Zhang H, Yuan Y. A Comparison of the Flavonoid Biosynthesis Mechanisms of Dendrobium Species by Analyzing the Transcriptome and Metabolome. International Journal of Molecular Sciences. 2022; 23(19):11980. https://doi.org/10.3390/ijms231911980
Chicago/Turabian StyleLiu, Sian, Hanyue Zhang, and Yingdan Yuan. 2022. "A Comparison of the Flavonoid Biosynthesis Mechanisms of Dendrobium Species by Analyzing the Transcriptome and Metabolome" International Journal of Molecular Sciences 23, no. 19: 11980. https://doi.org/10.3390/ijms231911980
APA StyleLiu, S., Zhang, H., & Yuan, Y. (2022). A Comparison of the Flavonoid Biosynthesis Mechanisms of Dendrobium Species by Analyzing the Transcriptome and Metabolome. International Journal of Molecular Sciences, 23(19), 11980. https://doi.org/10.3390/ijms231911980




