Deciphering the Host–Pathogen Interactome of the Wheat–Common Bunt System: A Step towards Enhanced Resilience in Next Generation Wheat
Abstract
:1. Introduction
2. Results and Discussion
2.1. HPIs from the Common Subnetwork (T. aestivum vs. T. caries/T. laevis)
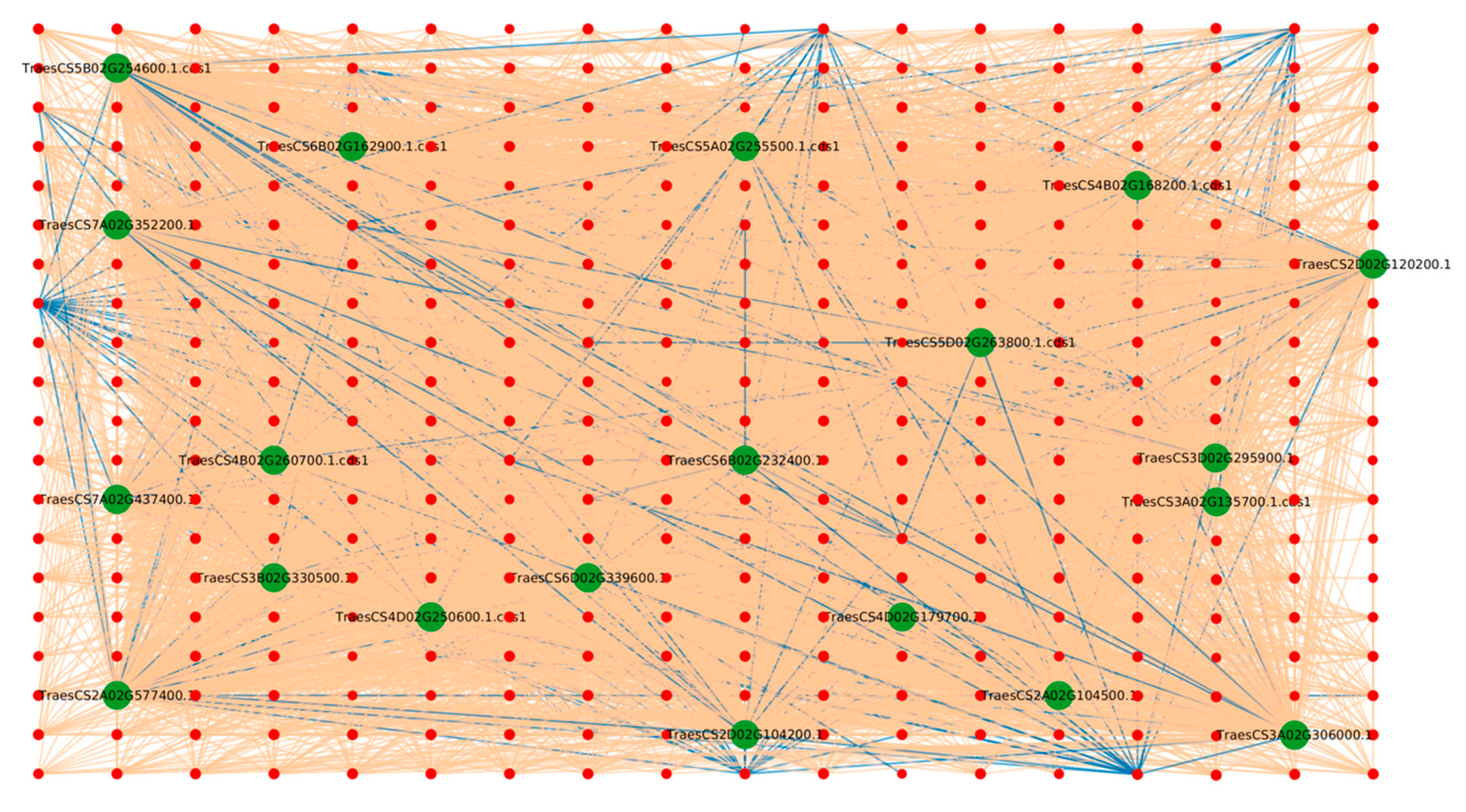
2.1.1. Protein Hubs Reveal the Major Proteins Involved in the Infection Mechanism
2.1.2. Degree
2.1.3. Tilletia Hubs
2.1.4. Triticum aestivum Hubs
2.2. GO Enrichment Analysis of the Proteins Involved in the Interactions
2.3. Analysis of Over-Represented KEGG Pathways
2.4. The Majority of Host–Pathogen Interactions Were Localized in the Plastid of Host Cells
2.5. Unique Interactions between Host and Pathogens
2.5.1. Functional Analysis of Unique T. caries Proteins in the Predicted PPIs
2.5.2. Functional Analysis of Unique T. laevis Proteins in the Predicted PPIs
2.6. Novel Host Targets Show Resistance to Common Bunt Disease
2.7. Identification of Stress-Related Transcription Factors in T. aestivum
3. Materials and Methods
3.1. Datasets
3.2. Prediction of PPIs between T. aestivum and Tilletia Species
3.2.1. Interolog-Based Prediction
3.2.2. Domain-Based Prediction
3.3. Prediction of Effector and Secretory Proteins
3.4. Functional Enrichment Analysis
3.5. Subcellular Localization of the Predicted Proteins
3.6. Comparison between HPIs of T. caries and T. laevis
3.7. Network Visualization
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miransari, M.; Smith, D. Sustainable wheat (Triticum aestivum L.) production in saline fields: A review. Crit. Rev. Biotechnol. 2019, 39, 999–1014. [Google Scholar] [CrossRef]
- Shewry, P.R.; Hey, S.J. The contribution of wheat to human diet and health. Food Energy Secur. 2015, 4, 178–202. [Google Scholar] [CrossRef]
- Narayanan, S. Effects of high temperature stress and traits associated with tolerance in wheat. Open Access J. Sci. 2018, 2, 177–186. [Google Scholar] [CrossRef]
- Figueroa, M.; Hammond-Kosack, K.E.; Solomon, P.S. A review of wheat diseases—A field perspective. Mol. Plant Pathol. 2018, 19, 1523–1536. [Google Scholar] [CrossRef]
- Dean, R.; Van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McNeil, M.; Roberts, A.M.I.; Cockerell, V.; Mulholland, V. Real-time PCR assay for quantification of Tilletia caries contamination of UK wheat seed. Plant Pathol. 2004, 53, 741–750. [Google Scholar] [CrossRef]
- Váňová, M.; Matušinsky, P.; Benada, J. Survey of Incidence of Bunts (Tilletia caries and Tilletia controversa) in the Czech Republic and Susceptibility of Winter Wheat Cultivars. Plant Prot. Sci. 2018, 42, 21–25. [Google Scholar] [CrossRef] [Green Version]
- Cota, L.; Botez, C.; Grigoras, M.; Curticiu, D. Screening for Resistance to Artificial Infection by Common Bunt (Tilletia caries and Tilletia Foetida) in F2 Populations of Wheat (Triticum aestivum L.). Bull. Univ. Agric. Sci. Vet. Med. Cluj-Napoca 2009, 66, 24–31. [Google Scholar]
- Li, C.; Wei, X.; Gao, L.; Chen, W.; Liu, T.; Liu, B. iTRAQ-Based Proteomic Analysis of Wheat Bunt Fungi Tilletia controversa, T. caries, and T. foetida. Curr. Microbiol. 2018, 75, 1103–1107. [Google Scholar] [CrossRef] [PubMed]
- Goates, B.J.; Bockelman, H.E. Identification of new sources of high levels of resistance to dwarf bunt and common bunt among winter wheat landraces in the USDA-ARS national small grains collection. Crop Sci. 2012, 52, 2595–2605. [Google Scholar] [CrossRef]
- Chen, J.; Guttieri, M.J.; Zhang, J.; Hole, D.; Souza, E.; Goates, B. A novel QTL associated with dwarf bunt resistance in Idaho 444 winter wheat. Theor. Appl. Genet. 2016, 129, 2313–2322. [Google Scholar] [CrossRef] [Green Version]
- Bonde, M.R.; Peterson, G.L.; Schaad, N.W.; Smilanick, J.L. Karnal bunt of wheat. Plant Dis. 1997, 81, 1370–1377. [Google Scholar] [CrossRef] [Green Version]
- Mourad, A.; Mahdy, E.; Bakheit, B.R.; Abo-elwafaa, A.; Baenziger, P.S. Effect of common bunt infection on agronomic traits in wheat (Triticum aestivum L.). J. Plant Genet. Breed. 2018, 2, 1–7. [Google Scholar]
- Pan, X.; Yang, Y.; Zhang, J.R. Molecular basis of host specificity in human pathogenic bacteria. Emerg. Microbes Infect. 2014, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, M.W.; Kann, M.G. Chapter 4: Protein Interactions and Disease. PLoS Comput. Biol. 2012, 8, e1002819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotlyar, M.; Pastrello, C.; Pivetta, F.; Lo Sardo, A.; Cumbaa, C.; Li, H.; Naranian, T.; Niu, Y.; Ding, Z.; Vafaee, F.; et al. In silico prediction of physical protein interactions and characterization of interactome orphans. Nat. Methods 2014, 12, 79–84. [Google Scholar] [CrossRef]
- Loaiza, C.D.; Duhan, N.; Lister, M.; Kaundal, R. In silico prediction of host–pathogen protein interactions in melioidosis pathogen Burkholderia pseudomallei and human reveals novel virulence factors and their targets. Brief. Bioinform. 2020, 22. [Google Scholar] [CrossRef]
- Wang, F.; Liu, M.; Song, B.; Li, D.; Pei, H.; Guo, Y.; Huang, J.; Zhang, D. Prediction and characterization of protein-protein interaction networks in swine. Proteome Sci. 2012, 10, 2. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.Y.; Deane, C.M.; Reinert, G. Predicting and validating protein interactions using network structure. PLoS Comput. Biol. 2008, 4, e1000118. [Google Scholar] [CrossRef] [Green Version]
- Ekman, D.; Light, S.; Björklund, Å.K.; Elofsson, A. What properties characterize the hub proteins of the protein-protein interaction network of Saccharomyces cerevisiae? Genome Biol. 2006, 7, R45. [Google Scholar] [CrossRef] [Green Version]
- Charitou, T.; Bryan, K.; Lynn, D.J. Using biological networks to integrate, visualize and analyze genomics data. Genet. Sel. Evol. 2016, 48, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, W.; Sang, C.; Shi, D.; Song, X.; Zhou, M.; Chen, C. Ubiquitin-like activating enzymes BcAtg3 and BcAtg7 participate in development and pathogenesis of Botrytis cinerea. Curr. Genet. 2018, 64, 919–930. [Google Scholar] [CrossRef] [PubMed]
- Dautt-Castro, M.; Rosendo-Vargas, M.; Casas-Flores, S. The Small GTPases in Fungal Signaling Conservation and Function. Cells 2021, 10, 1039. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Yao, J.; Zhang, Y.; Li, S.; Kang, Z. Characterization of a Ran gene from Puccinia striiformis f. sp. tritici involved in fungal growth and anti-cell death. Sci. Rep. 2016, 6, 35248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heupel, S.; Roser, B.; Kuhn, H.; Lebrun, M.H.; Villalba, F.; Requena, N. Erl1, a novel era-like GTPase from Magnaporthe oryzae, is required for full root virulence and is conserved in the mutualistic symbiont Glomus intraradices. Mol. Plant Microbe Interact. 2010, 23, 67–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Li, G.; Zhang, S.; Jiang, C.; Qin, J.; Xu, J.R. Activation of the signalling mucin MoMsb2 and its functional relationship with Cbp1 in Magnaporthe oryzae. Environ. Microbiol. 2015, 17, 2969–2981. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.J.; Kim, K.T.; Lee, Y.H. SUMOylation is required for fungal development and pathogenicity in the rice blast fungus Magnaporthe oryzae. Mol. Plant Pathol. 2018, 19, 2134–2148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, X.; He, F.; Ding, M.; Geng, C.; Chen, L.; Zou, S.; Liang, Y.; Yu, J.; Dong, H. Thioredoxin reductase is involved in development and pathogenicity in Fusarium graminearum. Front. Microbiol. 2019, 10, 393. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Cui, W.; Abdul Haseeb, H.; Guo, W. VdNop12, containing two tandem RNA recognition motif domains, is a crucial factor for pathogenicity and cold adaption in Verticillium dahliae. Environ. Microbiol. 2020, 22, 5387–5401. [Google Scholar] [CrossRef] [PubMed]
- Becht, P.; Vollmeister, E.; Feldbrügge, M. Role for RNA-binding proteins implicated in pathogenic development of Ustilago maydis. Eukaryot. Cell 2005, 4, 121–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Yan, H.; Qiu, Z.; Hu, B.; Zeng, B.; Zhong, C.; Fan, C. Comprehensive analysis of SNRK gene family and their responses to salt stress in Eucalyptus grandis. Int. J. Mol. Sci. 2019, 20, 2786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Loh, Y.T.; Bressan, R.A.; Martin, G.B. The tomato gene Pti1 encodes a serine/threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive response. Cell 1995, 83, 925–935. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Yun, H.S.; Kwon, C. Molecular communications between plant heat shock responses and disease resistance. Mol. Cells 2012, 34, 109–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alam, S.B.; Rochon, D. Cucumber Necrosis Virus Recruits Cellular Heat Shock Protein 70 Homologs at Several Stages of Infection. J. Virol. 2016, 90, 3302–3317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, H.L. Functional Roles of Plant Protein Kinases in Signal Transduction Pathways during Abiotic and Biotic Stress. J. Biodivers. Bioprospecting Dev. 2015, 2. [Google Scholar] [CrossRef]
- Asano, T.; Nguyen, T.H.N.; Yasuda, M.; Sidiq, Y.; Nishimura, K.; Nakashita, H.; Nishiuchi, T. Arabidopsis MAPKKK δ-1 is required for full immunity against bacterial and fungal infection. J. Exp. Bot. 2020, 71, 2085–2097. [Google Scholar] [CrossRef] [PubMed]
- Shou, H.; Bordallo, P.; Wang, K. Expression of the Nicotiana protein kinase (NPK1) enhanced drought tolerance in transgenic maize. J. Exp. Bot. 2004, 55, 1013–1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovtun, Y.; Chiu, W.L.; Tena, G.; Sheen, J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA 2000, 97, 2940–2945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, X.; Gai, W.X.; Qiao, Y.M.; Ali, M.; Wei, A.M.; Luo, D.X.; Li, Q.H.; Gong, Z.H. Identification of CBL and CIPK gene families and functional characterization of CaCIPK1 under Phytophthora capsici in pepper (Capsicum annuum L.). BMC Genom. 2019, 20, 775. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Guo, J.; Zhang, R.; Zhao, J.; Liu, C.; Qi, T.; Duan, Y.; Kang, Z.; Guo, J. TaCIPK10 interacts with and phosphorylates TaNH2 to activate wheat defense responses to stripe rust. Plant Biotechnol. J. 2019, 17, 956–968. [Google Scholar] [CrossRef]
- Zhu, X.; Yang, K.; Wei, X.; Zhang, Q.; Rong, W.; Du, L.; Ye, X.; Qi, L.; Zhang, Z. The wheat AGC kinase TaAGC1 is a positive contributor to host resistance to the necrotrophic pathogen Rhizoctonia cerealis. J. Exp. Bot. 2015, 66, 6591–6603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinter, N.; Hach, C.A.; Hampel, M.; Rekhter, D.; Zienkiewicz, K.; Feussner, I.; Poehlein, A.; Daniel, R.; Finkernagel, F.; Heimel, K. Signal peptide peptidase activity connects the unfolded protein response to plant defense suppression by Ustilago maydis. PloS Pathog. 2019, 15, e1007734. [Google Scholar] [CrossRef] [PubMed]
- Pogány, M.; Dankó, T.; Kámán-Tóth, E.; Schwarczinger, I.; Bozsó, Z. Regulatory proteolysis in Arabidopsis-Pathogen interactions. Int. J. Mol. Sci. 2015, 16, 23177–23194. [Google Scholar] [CrossRef] [Green Version]
- Devoto, A.; Muskett, P.R.; Shirasu, K. Role of ubiquitination in the regulation of plant defence against pathogens. Curr. Opin. Plant Biol. 2003, 6, 307–311. [Google Scholar] [CrossRef]
- Wang, S.; Cao, L.; Wang, H. Arabidopsis ubiquitin-conjugating enzyme UBC22 is required for female gametophyte development and likely involved in Lys11-linked ubiquitination. J. Exp. Bot. 2016, 67, 3277–3288. [Google Scholar] [CrossRef] [Green Version]
- Balasubramanian, V.; Vashisht, D.; Cletus, J.; Sakthivel, N. Plant β-1,3-glucanases: Their biological functions and transgenic expression against phytopathogenic fungi. Biotechnol. Lett. 2012, 34, 1983–1990. [Google Scholar] [CrossRef]
- Aggarwal, R.; Purwar, S.; Kharbikar, L.; Gupta, S. Induction of a wheat β-1,3-glucanase gene during the defense response to Bipolaris sorokiniana. Acta Phytopathol. Entomol. Hung. 2011, 46, 39–47. [Google Scholar] [CrossRef]
- Breuers, F.K.H.; Bräutigam, A.; Weber, A.P.M. The plastid outer envelope—A highly dynamic interface between plastid and cytoplasm. Front. Plant Sci. 2011, 2, 97. [Google Scholar] [CrossRef] [Green Version]
- Howe, G.A.; Schilmiller, A.L. Oxylipin metabolism in response to stress. Curr. Opin. Plant Biol. 2002, 5, 230–236. [Google Scholar] [CrossRef]
- Kishimoto, K.; Matsui, K.; Ozawa, R.; Takabayashi, J. Direct fungicidal activities of C6-aldehydes are important constituents for defense responses in Arabidopsis against Botrytis cinerea. Phytochemistry 2008, 69, 2127–2132. [Google Scholar] [CrossRef]
- Ferro, M.; Salvi, D.; Brugière, S.; Miras, S.; Kowalski, S.; Louwagie, M.; Garin, J.; Joyard, J.; Rolland, N. Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol. Cell. Proteom. 2003, 2, 325–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fritsche, S.; Wang, X.; Jung, C. Recent advances in our understanding of tocopherol biosynthesis in plants: An overview of key genes, functions, and breeding of vitamin E improved crops. Antioxidants 2017, 6, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nosek, M.; Kornaś, A.; Kuźniak, E.; Miszalski, Z. Plastoquinone redox state modifies plant response to pathogen. Plant Physiol. Biochem. 2015, 96, 163–170. [Google Scholar] [CrossRef]
- Liu, Z.; Fan, M.; Li, C.; Xu, J.H. Dynamic gene amplification and function diversification of grass-specific O-methyltransferase gene family. Genomics 2019, 111, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Bhuiyan, N.H.; Selvaraj, G.; Wei, Y.; King, J. Gene expression profiling and silencing reveal that monolignol biosynthesis plays a critical role in penetration defence in wheat against powdery mildew invasion. J. Exp. Bot. 2009, 60, 509–521. [Google Scholar] [CrossRef]
- Wang, M.; Zhu, X.; Wang, K.; Lu, C.; Luo, M.; Shan, T.; Zhang, Z. A wheat caffeic acid 3-O-methyltransferase TaCOMT-3D positively contributes to both resistance to sharp eyespot disease and stem mechanical strength. Sci. Rep. 2018, 8, 6543. [Google Scholar] [CrossRef] [PubMed]
- Heyno, E.; Alkan, N.; Fluhr, R. A dual role for plant quinone reductases in host-fungus interaction. Physiol. Plant. 2013, 149, 340–353. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Joy, J.; Zhou, W.; De, S.; Wood, W.H.; Becker, K.G.; Ji, H.; Sen, R. Transcriptional outcomes and kinetic patterning of gene expression in response to NF-κB activation. PLoS Biol. 2018, 16, e2006347. [Google Scholar] [CrossRef] [Green Version]
- Ryals, J.; Weymann, K.; Lawton, K.; Friedrich, L.; Ellis, D.; Steiner, H.Y.; Johnson, J.; Delaney, T.P.; Jesse, T.; Vos, P.; et al. The arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor IκB. Plant Cell 1997, 9, 425–439. [Google Scholar] [CrossRef] [Green Version]
- Hiscott, J.; Nguyen, T.L.A.; Arguello, M.; Nakhaei, P.; Paz, S. Manipulation of the nuclear factor-κB pathway and the innate immune response by viruses. Oncogene 2006, 25, 6844–6867. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Klessig, D.F. MAPK cascades in plant defense signaling. Trends Plant Sci. 2001, 6, 520–527. [Google Scholar] [CrossRef]
- Jalmi, S.K.; Sinha, A.K. Functional Involvement of a Mitogen Activated Protein Kinase Module, OsMKK3-OsMPK7-OsWRK30 in Mediating Resistance against Xanthomonas oryzae in Rice. Sci. Rep. 2016, 6, 37974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drawid, A.; Gerstein, M. A Bayesian system integrating expression data with sequence patterns for localizing proteins: Comprehensive application to the yeast genome. J. Mol. Biol. 2000, 301, 1059–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Li, C.; Wang, E.; Wang, X. An FPT approach for predicting protein localization from yeast genomic data. PLoS ONE 2011, 6, e14449. [Google Scholar] [CrossRef]
- Pedrajas, J.R.; Kosmidou, E.; Miranda-Vizuete, A.; Gustafsson, J.Å.; Wright, A.P.H.; Spyrou, G. Identification and functional characterization of a novel mitochondrial thioredoxin system in Saccharomyces cerevisiae. J. Biol. Chem. 1999, 274, 6366–6373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trotter, E.W.; Grant, C.M. Overlapping roles of the cytoplasmic and mitochondrial redox regulatory systems in the yeast Saccharomyces cerevisiae. Eukaryot. Cell 2005, 4, 392–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.Y.; Kwon, S.I.; Choi, C.; Lee, H.; Ahn, I.; Park, S.R.; Bae, S.C.; Lee, S.C.; Hwang, D.J. Expression analysis of rice VQ genes in response to biotic and abiotic stresses. Gene 2013, 529, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Huberts, D.H.E.W.; van der Klei, I.J. Moonlighting proteins: An intriguing mode of multitasking. Biochim. Biophys. Acta Mol. Cell Res. 2010, 1803, 520–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, T.; Zhao, J.; Ying, S.; Feng, M. Differential Contributions of Five ABC Transporters to Mutidrug Resistance, Antioxidion and Virulence of Beauveria bassiana, an Entomopathogenic Fungus. PLoS ONE 2013, 8, e62179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Floryszak-wieczorek, J.; Gajewska, J.; Gzyl, J.; Jelonek, T.; Arasimowicz-jelonek, M. Switchable Nitroproteome States of Phytophthora infestans Biology and Pathobiology. Front. Microbiol. 2019, 10, 1516. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Elfstrand, M.; Stenlid, J.; Durling, M.B.; Olson, Å. The conifer root rot pathogens Heterobasidion irregulare and Heterobasidion occidentale employ different strategies to infect Norway spruce. Sci. Rep. 2020, 10, 5884. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Wei, S.; Yang, X.; Liu, W. Proteomic analysis of exudate of Cercospora armoraciae from Armoracia rusticana. PeerJ 2020, 8, e9592. [Google Scholar] [CrossRef] [PubMed]
- Andersen, E.; Lindsey, L.; Nepal, M. Genome-wide Identification of Disease Resistance Genes (R Genes) in Wheat. bioRxiv 2020. [Google Scholar] [CrossRef]
- Bhatta, M.; Morgounov, A.; Belamkar, V.; Yorgancılar, A.; Baenziger, P.S. Genome-wide association study reveals favorable alleles associated with common bunt resistance in synthetic hexaploid wheat. Euphytica 2018, 214, 200. [Google Scholar] [CrossRef]
- Mourad, A.M.I.; Sallam, A.; Belamkar, V.; Mahdy, E.; Bakheit, B.; Abo El-Wafaa, A.; Stephen Baenziger, P. Genetic architecture of common bunt resistance in winter wheat using genome-wide association study. BMC Plant Biol. 2018, 18, 280. [Google Scholar] [CrossRef]
- Muellner, A.E.; Buerstmayr, M.; Eshonkulov, B.; Hole, D.; Michel, S.; Hagenguth, J.F.; Pachler, B.; Pernold, R.; Buerstmayr, H. Comparative mapping and validation of multiple disease resistance QTL for simultaneously controlling common and dwarf bunt in bread wheat. Theor. Appl. Genet. 2021, 134, 489–503. [Google Scholar] [CrossRef] [PubMed]
- Hafeez, A.N.; Arora, S.; Ghosh, S.; Gilbert, D.; Bowden, R.L.; Wulff, B.B.H. Creation and judicious application of a wheat resistance gene atlas. Mol. Plant 2021, 14, 1053–1070. [Google Scholar] [CrossRef]
- Baillo, E.H.; Kimotho, R.N.; Zhang, Z.; Xu, P. Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes 2019, 10, 771. [Google Scholar] [CrossRef] [Green Version]
- Ng, D.W.K.; Abeysinghe, J.K.; Kamali, M. Regulating the regulators: The control of transcription factors in plant defense signaling. Int. J. Mol. Sci. 2018, 19, 3737. [Google Scholar] [CrossRef] [Green Version]
- Yuan, X.; Wang, H.; Cai, J.; Li, D.; Song, F. NAC transcription factors in plant immunity. Phytopathol. Res. 2019, 1, 3. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Kerrien, S.; Aranda, B.; Breuza, L.; Bridge, A.; Broackes-Carter, F.; Chen, C.; Duesbury, M.; Dumousseau, M.; Feuermann, M.; Hinz, U.; et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012, 40, 841–846. [Google Scholar] [CrossRef] [PubMed]
- Licata, L.; Briganti, L.; Peluso, D.; Perfetto, L.; Iannuccelli, M.; Galeota, E.; Sacco, F.; Palma, A.; Nardozza, A.P.; Santonico, E.; et al. MINT, the molecular interaction database: 2012 Update. Nucleic Acids Res. 2012, 40, 857–861. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Nanduri, B. HPIDB—A unified resource for host-pathogen interactions. BMC Bioinform. 2010, 11, 16. [Google Scholar] [CrossRef] [Green Version]
- Salwinski, L.; Miller, C.S.; Smith, A.J.; Pettit, F.K.; Bowie, J.U.; Eisenberg, D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004, 32, 449–451. [Google Scholar] [CrossRef] [Green Version]
- Chatr-Aryamontri, A.; Oughtred, R.; Boucher, L.; Rust, J.; Chang, C.; Kolas, N.K.; O’Donnell, L.; Oster, S.; Theesfeld, C.; Sellam, A.; et al. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017, 45, D369–D379. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Urban, M.; Cuzick, A.; Seager, J.; Wood, V.; Rutherford, K.; Venkatesh, S.Y.; De Silva, N.; Martinez, M.C.; Pedro, H.; Yates, A.D.; et al. PHI-base: The pathogen-host interactions database. Nucleic Acids Res. 2020, 48, D613–D620. [Google Scholar] [CrossRef]
- Mosca, R.; Céol, A.; Stein, A.; Olivella, R.; Aloy, P. 3did: A catalog of domain-based interactions of known three-dimensional structure. Nucleic Acids Res. 2014, 42, 374–379. [Google Scholar] [CrossRef] [Green Version]
- Raghavachari, B.; Tasneem, A.; Przytycka, T.M.; Jothi, R. DOMINE: A database of protein domain interactions. Nucleic Acids Res. 2008, 36, 656–661. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Min, B.; Yi, G.S. IDDI: Integrated domain-domain interaction and protein interaction analysis system. Proteome Sci. 2012, 10, S9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, 222–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nourani, E.; Khunjush, F.; Durmus, S. Computational approaches for prediction of pathogen-host protein-protein interactions. Front. Microbiol. 2015, 6, 94. [Google Scholar] [CrossRef]
- Huo, T.; Liu, W.; Guo, Y.; Yang, C.; Lin, J.; Rao, Z. Prediction of host-pathogen protein interactions between Mycobacterium tuberculosis and Homo sapiens using sequence motifs. BMC Bioinform. 2015, 16, 100. [Google Scholar] [CrossRef] [Green Version]
- Thanasomboon, R.; Kalapanulak, S.; Netrphan, S.; Saithong, T. Exploring dynamic protein-protein interactions in cassava through the integrative interactome network. Sci. Rep. 2020, 10, 6510. [Google Scholar] [CrossRef] [PubMed]
- Wojcik, J.; Schächter, V. Protein-protein interaction map inference using interacting domain profile pairs. Bioinformatics 2001, 17 (Suppl. S1), S296–S305. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S.I.; Mahmud, Z.; Elahi, M.; Akter, A.; Jewel, N.A.; Muzahidul Islam, M.; Ferdous, S.; Kikuchi, T. Study of intra–inter species protein–protein interactions for potential drug targets identification and subsequent drug design for Escherichia coli O104:H4 C277-11. In Silico Pharmacol. 2017, 5, 1. [Google Scholar] [CrossRef] [Green Version]
- Lian, X.; Yang, X.; Shao, J.; Hou, F.; Yang, S.; Pan, D.; Zhang, Z. Prediction and analysis of human-herpes simplex virus type 1 protein-protein interactions by integrating multiple methods. Quant. Biol. 2020, 8, 312–324. [Google Scholar] [CrossRef]
- Sperschneider, J.; Dodds, P.N.; Gardiner, D.M.; Singh, K.B.; Taylor, J.M. Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Mol. Plant Pathol. 2018, 19, 2094–2110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feldman, D.; Yarden, O.; Hadar, Y. Seeking the Roles for Fungal Small-Secreted Proteins in Affecting Saprophytic Lifestyles. Front. Microbiol. 2020, 11, 455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Dönnes, P.; Höglund, A. Predicting protein subcellular localization: Past, present, and future. Genom. Proteom. Bioinform. 2004, 2, 209–215. [Google Scholar] [CrossRef] [Green Version]
- Sperschneider, J.; Catanzariti, A.M.; Deboer, K.; Petre, B.; Gardiner, D.M.; Singh, K.B.; Dodds, P.N.; Taylor, J.M. LOCALIZER: Subcellular localization prediction of both plant and effector proteins in the plant cell. Sci. Rep. 2017, 7, rep44598. [Google Scholar] [CrossRef] [Green Version]
- Sahu, S.S.; Loaiza, C.D.; Kaundal, R. Plant-mSubP: A computational framework for the prediction of single- and multi-target protein subcellular localization using integrated machine-learning approaches. AoB Plants 2020, 12, plz068. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Sønderby, C.K.; Sønderby, S.K.; Nielsen, H.; Winther, O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef] [PubMed]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramos, P.I.P.; Arge, L.W.P.; Lima, N.C.B.; Fukutani, K.F.; de Queiroz, A.T.L. Leveraging User-Friendly Network Approaches to Extract Knowledge from High-Throughput Omics Datasets. Front. Genet. 2019, 10, 1120. [Google Scholar] [CrossRef]
- Safari-Alighiarloo, N.; Taghizadeh, M.; Rezaei-Tavirani, M.; Goliaei, B.; Peyvandi, A.A. Protein-protein interaction networks (PPI) and complex diseases. Gastroenterol. Hepatol. Bed Bench 2014, 7, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]








| Interaction Database | Number of Interactions | Number of Host Proteins | Number of Pathogen Proteins |
|---|---|---|---|
| Interolog-based | |||
| BioGRID | 11,343,237 | 51,839 | 3574 |
| DIP | 1,334,228 | 27,027 | 2404 |
| HPIDB | 48,331 | 6779 | 247 |
| IntAct | 4,768,852 | 48,915 | 3320 |
| MINT | 1,338,779 | 23,156 | 2608 |
| PHI-base | 28 | 7 | 4 |
| STRING | 31,159,410 | 82,876 | 2638 |
| Total (Interolog) (I) | 37,714,442 | 83,639 | 3872 |
| Domain-based | |||
| 3DID | 1,221,946 | 27,342 | 2612 |
| DOMINE | 5,803,329 | 28,007 | 2623 |
| IDDI | 12,112,523 | 33,619 | 3235 |
| Total (Domain) (II) | 14,307,366 | 35,526 | 3396 |
| I and II (combined) | 46,557,278 | 83,947 | 4612 |
| I and II (consensus) | 5,464,530 | 30,629 | 2401 |
| Interolog (unique) | 32,249,912 | 83,637 | 3867 |
| Domain (unique) | 8,842,836 | 34,390 | 3348 |
| Interaction Database | Number of Interactions | Number of Host Proteins | Number of Pathogen Proteins |
|---|---|---|---|
| Interolog-based | |||
| BioGRID | 11,003,345 | 51,985 | 3417 |
| DIP | 1,263,028 | 26,726 | 2307 |
| HPIDB | 46,117 | 6669 | 227 |
| IntAct | 4,601,434 | 48,463 | 3183 |
| MINT | 1,278,978 | 22,970 | 2536 |
| PHI-base | 35 | 7 | 5 |
| STRING | 29,978,511 | 82,878 | 2558 |
| Total (Interolog) (I) | 36,330,023 | 83,637 | 3697 |
| Domain-based | |||
| 3DID | 1,151,885 | 27,110 | 2483 |
| DOMINE | 5,491,200 | 27,901 | 2502 |
| IDDI | 11,548,603 | 33,704 | 3063 |
| Total (Domain) (II) | 13,642,742 | 35,591 | 3212 |
| I and II (combined) | 44,725,200 | 83,941 | 4380 |
| I and II (consensus) | 5,247,565 | 30,659 | 2305 |
| Interolog (unique) | 31,082,458 | 83,634 | 3692 |
| Domain (unique) | 8,395,177 | 34,176 | 3164 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kataria, R.; Kaundal, R. Deciphering the Host–Pathogen Interactome of the Wheat–Common Bunt System: A Step towards Enhanced Resilience in Next Generation Wheat. Int. J. Mol. Sci. 2022, 23, 2589. https://doi.org/10.3390/ijms23052589
Kataria R, Kaundal R. Deciphering the Host–Pathogen Interactome of the Wheat–Common Bunt System: A Step towards Enhanced Resilience in Next Generation Wheat. International Journal of Molecular Sciences. 2022; 23(5):2589. https://doi.org/10.3390/ijms23052589
Chicago/Turabian StyleKataria, Raghav, and Rakesh Kaundal. 2022. "Deciphering the Host–Pathogen Interactome of the Wheat–Common Bunt System: A Step towards Enhanced Resilience in Next Generation Wheat" International Journal of Molecular Sciences 23, no. 5: 2589. https://doi.org/10.3390/ijms23052589
APA StyleKataria, R., & Kaundal, R. (2022). Deciphering the Host–Pathogen Interactome of the Wheat–Common Bunt System: A Step towards Enhanced Resilience in Next Generation Wheat. International Journal of Molecular Sciences, 23(5), 2589. https://doi.org/10.3390/ijms23052589







