The Complex Proteolipidic Behavior of the SARS-CoV-2 Envelope Protein Channel: Weak Selectivity and Heterogeneous Oligomerization
Abstract
:1. Introduction
2. Results
2.1. SARS-2 E Is a Voltage-Dependent Ohmic Channel That Is Not Activated by Voltage
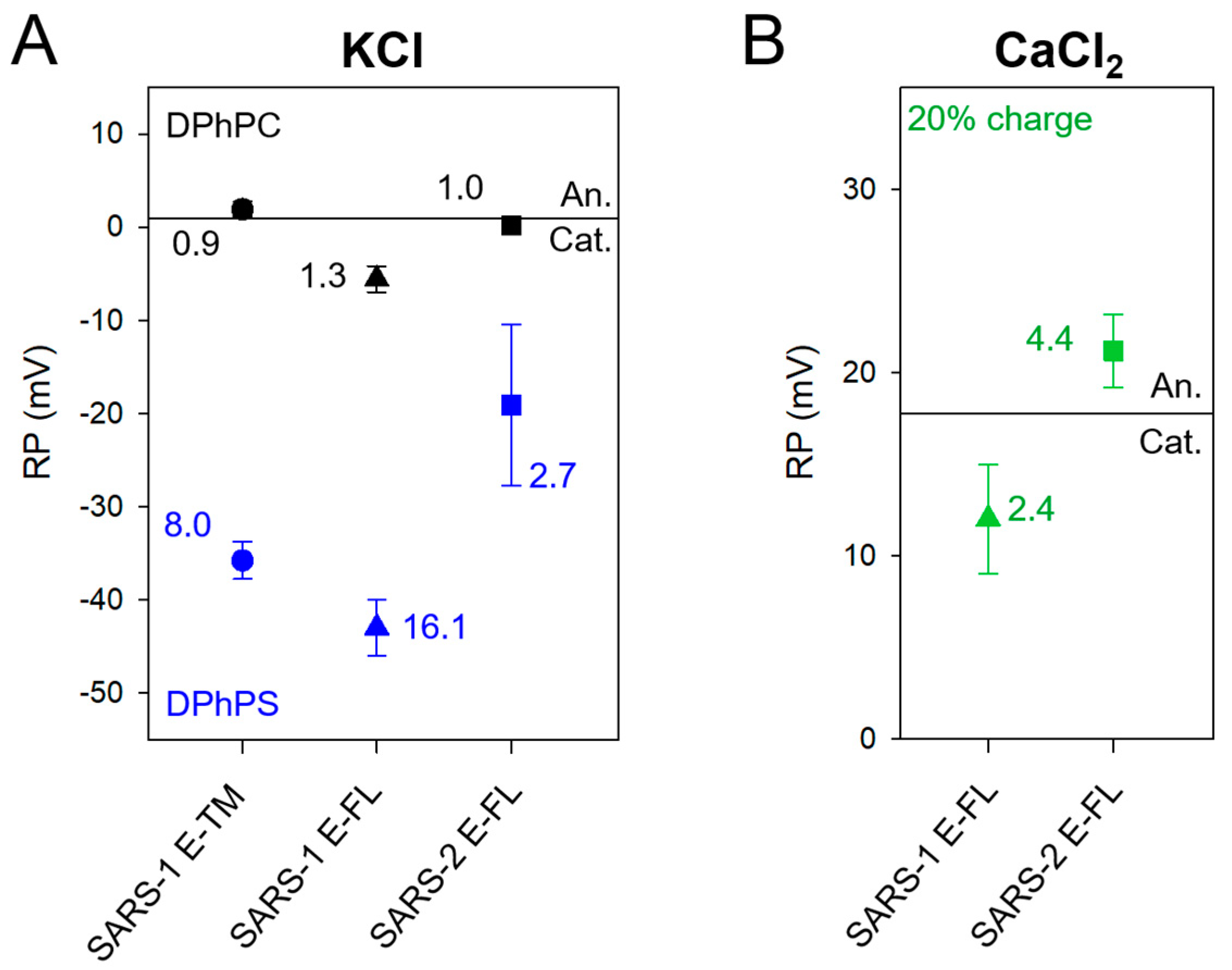
2.2. SARS-2 E Channels Are Mildly Selective but Strongly Modulated by Lipid Charge
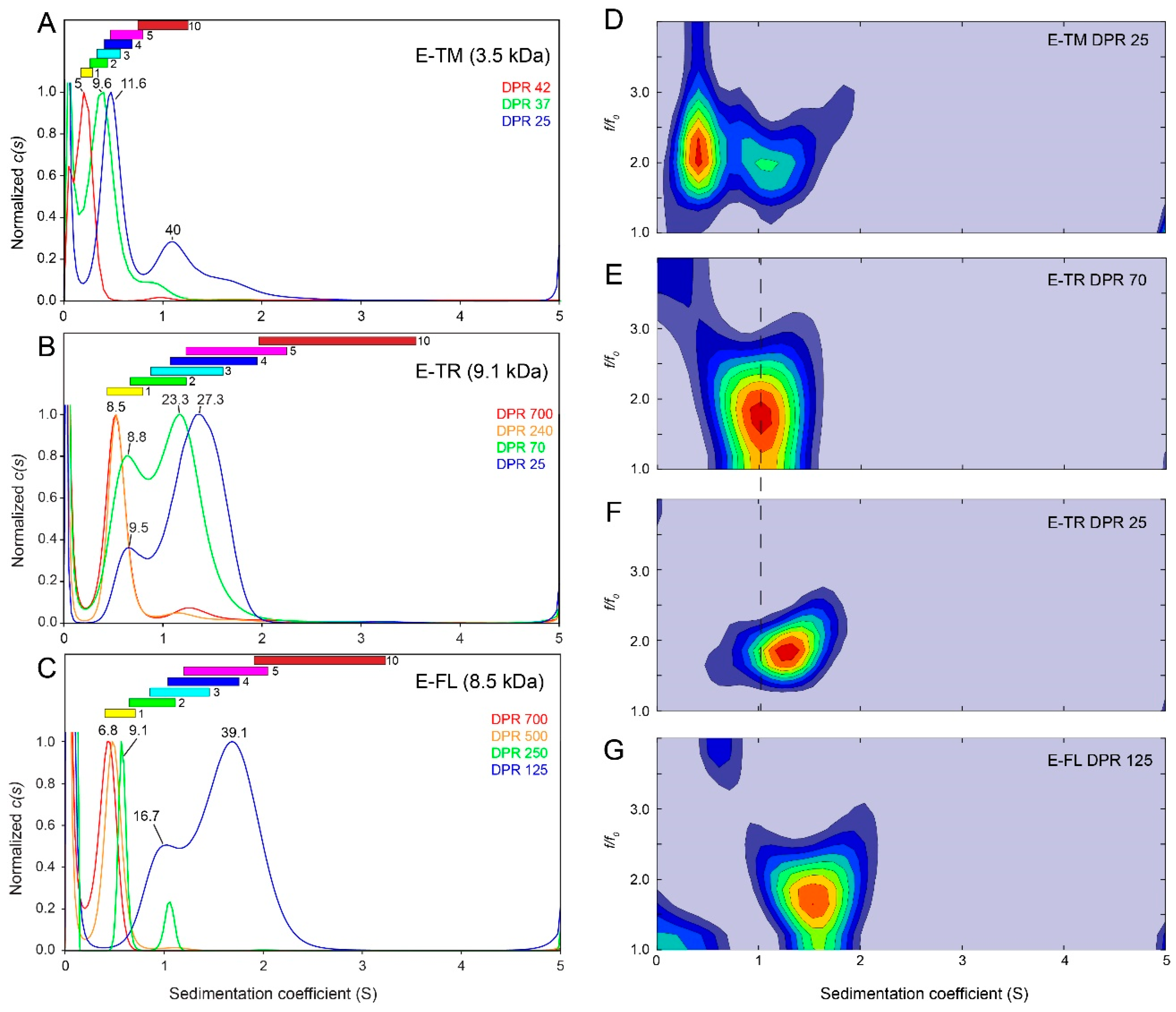
2.3. SARS-2 E Oligomerization in Detergent
3. Discussion
4. Materials and Methods
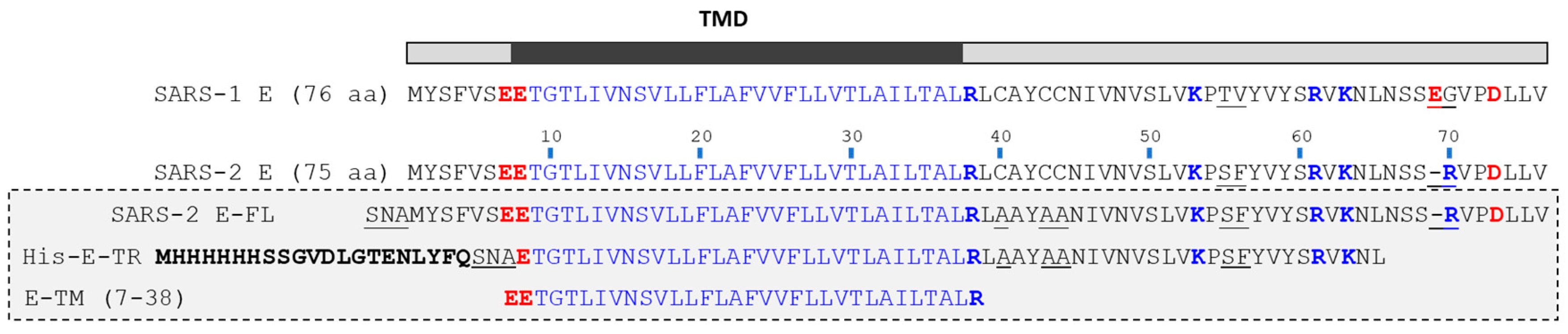
4.1. E Protein Synthesis, Expression, and Purification
4.2. Analytical Ultracentrifugation Sedimentation Velocity (AUC-SV)
4.3. Planar Membrane Formation and Ion Channel Reconstitution
4.4. Ionic Current Recording and Selectivity Measurements
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- To, J.; Surya, W.; Torres, J. Targeting the Channel Activity of Viroporins. Adv. Protein Chem. Struct. Biol. 2016, 104, 307–355. [Google Scholar] [CrossRef]
- Nieto-Torres, J.L.; DeDiego, M.L.; Álvarez, E.; Jiménez-Guardeño, J.M.; Regla-Nava, J.A.; Llorente, M.; Kremer, L.; Shuo, S.; Enjuanes, L. Subcellular Location and Topology of Severe Acute Respiratory Syndrome Coronavirus Envelope Protein. Virology 2011, 415, 69–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandala, V.S.; McKay, M.J.; Shcherbakov, A.A.; Dregni, A.J.; Kolocouris, A.; Hong, M. Structure and Drug Binding of the SARS-CoV-2 Envelope Protein Transmembrane Domain in Lipid Bilayers. Nat. Struct. Mol. Biol. 2020, 27, 1202–1208. [Google Scholar] [CrossRef]
- Torres, J.; Maheswari, U.; Parthasarathy, K.; Ng, L.; Liu, D.X.; Gong, X. Conductance and Amantadine Binding of a Pore Formed by a Lysine-Flanked Transmembrane Domain of SARS Coronavirus Envelope Protein. Protein Sci. 2007, 16, 2065–2071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, L.; Mckinlay, C.; Gage, P.; Ewart, G. SARS Coronavirus E Protein Forms Cation-Selective Ion Channels. Virology 2004, 330, 322–331. [Google Scholar] [CrossRef] [Green Version]
- Nieto-Torres, J.L.; DeDiego, M.L.; Verdiá-Báguena, C.; Jimenez-Guardeño, J.M.; Regla-Nava, J.A.; Fernandez-Delgado, R.; Castaño-Rodriguez, C.; Alcaraz, A.; Torres, J.; Aguilella, V.M.; et al. Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Ion Channel Activity Promotes Virus Fitness and Pathogenesis. PLoS Pathog. 2014, 10, e1004077. [Google Scholar] [CrossRef] [Green Version]
- Parthasarathy, K.; Lu, H.; Surya, W.; Vararattanavech, A.; Pervushin, K.; Torres, J. Expression and Purification of Coronavirus Envelope Proteins Using a Modified β-Barrel Construct. Protein Expr. Purif. 2012, 85, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Parthasarathy, K.; Ng, L.; Lin, X.; Liu, D.X.; Pervushin, K.; Gong, X.; Torres, J. Structural Flexibility of the Pentameric SARS Coronavirus Envelope Protein Ion Channel. Biophys. J. 2008, 95, L39–L41. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Surya, W.; Claudine, S.; Torres, J. Structure of a Conserved Golgi Complex-Targeting Signal in Coronavirus Envelope Proteins. J. Biol. Chem. 2014, 289, 12535–12549. [Google Scholar] [CrossRef] [Green Version]
- Nieto-Torres, J.L.; Verdiá-Báguena, C.; Jimenez-Guardeño, J.M.; Regla-Nava, J.A.; Castaño-Rodriguez, C.; Fernandez-Delgado, R.; Torres, J.; Aguilella, V.M.; Enjuanes, L. Severe Acute Respiratory Syndrome Coronavirus E Protein Transports Calcium Ions and Activates the NLRP3 Inflammasome. Virology 2015, 485, 330–339. [Google Scholar] [CrossRef] [Green Version]
- Verdiá-Báguena, C.; Nieto-Torres, J.L.; Alcaraz, A.; DeDiego, M.L.; Torres, J.; Aguilella, V.M.; Enjuanes, L. Coronavirus E Protein Forms Ion Channels with Functionally and Structurally-Involved Membrane Lipids. Virology 2012, 432, 485–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verdiá-Báguena, C.; Nieto-Torres, J.L.; Alcaraz, A.; DeDiego, M.L.; Enjuanes, L.; Aguilella, V.M. Analysis of SARS-CoV E Protein Ion Channel Activity by Tuning the Protein and Lipid Charge. Biochim. Et Biophys. Acta (BBA)—Biomembr. 2013, 1828, 2026–2031. [Google Scholar] [CrossRef] [Green Version]
- Somberg, N.H.; Wu, W.W.; Medeiros-Silva, J.; Dregni, A.J.; Jo, H.; Degrado, W.F.; Hong, M. SARS-CoV-2 Envelope Protein Forms Clustered Pentamers in Lipid Bilayers. Biochemistry 2022, 61, 2280–2294. [Google Scholar] [CrossRef] [PubMed]
- Surya, W.; Torres, J. Oligomerization-Dependent Beta-Structure Formation in SARS-CoV-2 Envelope Protein. Int. J. Mol. Sci. 2022, 23, 13285. [Google Scholar] [CrossRef] [PubMed]
- Poggio, E.; Vallese, F.; Hartel, A.J.W.; Morgenstern, T.J.; Kanner, S.A.; Rauh, O.; Giamogante, F.; Barazzuol, L.; Shepard, K.L.; Colecraft, H.M.; et al. Perturbation of the Host Cell Ca2+ Homeostasis and ER-Mitochondria Contact Sites by the SARS-CoV-2 Structural Proteins E and M. Cell Death Dis. 2023, 14, 297. [Google Scholar] [CrossRef] [PubMed]
- Pervushin, K.; Tan, E.; Parthasarathy, K.; Lin, X.; Jiang, F.L.; Yu, D.; Vararattanavech, A.; Tuck, W.S.; Ding, X.L.; Torres, J. Structure and Inhibition of the SARS Coronavirus Envelope Protein Ion Channel. PLoS Pathog. 2009, 5, e1000511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torres, J.; Parthasarathy, K.; Lin, X.; Saravanan, R.; Kukol, A.; Liu, D.X. Model of a Putative Pore: The Pentameric Alpha-Helical Bundle of SARS Coronavirus E Protein in Lipid Bilayers. Biophys. J. 2006, 91, 938–947. [Google Scholar] [CrossRef] [Green Version]
- Medeiros-Silva, J.; Somberg, N.H.; Wang, H.K.; McKay, M.J.; Mandala, V.S.; Dregni, A.J.; Hong, M. PH- and Calcium-Dependent Aromatic Network in the SARS-CoV-2 Envelope Protein. J. Am. Chem. Soc. 2022, 144, jacs.2c00973. [Google Scholar] [CrossRef]
- Surya, W.; Li, Y.; Torres, J. Structural Model of the SARS Coronavirus E Channel in LMPG Micelles. Biochim. Biophys. Acta Biomembr. 2018, 1860, 1309–1317. [Google Scholar] [CrossRef]
- Park, S.H.; Siddiqi, H.; Castro, D.V.; De Angelis, A.A.; Oom, A.L.; Stoneham, C.A.; Lewinski, M.K.; Clark, A.E.; Croker, B.A.; Carlin, A.F.; et al. Interactions of SARS-CoV-2 Envelope Protein with Amilorides Correlate with Antiviral Activity. PLoS Pathog. 2021, 17, e1009519. [Google Scholar] [CrossRef]
- Dregni, A.J.; McKay, M.J.; Surya, W.; Queralt-Martin, M.; Medeiros-Silva, J.; Wang, H.K.; Aguilella, V.; Torres, J.; Hong, M. The Cytoplasmic Domain of the SARS-CoV-2 Envelope Protein Assembles into a β-Sheet Bundle in Lipid Bilayers. J. Mol. Biol. 2023, 435, 167966. [Google Scholar] [CrossRef]
- Verdiá-Báguena, C.; Aguilella, V.M.; Queralt-Martín, M.; Alcaraz, A. Transport Mechanisms of SARS-CoV-E Viroporin in Calcium Solutions: Lipid-Dependent Anomalous Mole Fraction Effect and Regulation of Pore Conductance. Biochim. Biophys. Acta Biomembr. 2021, 1863, 183590. [Google Scholar] [CrossRef]
- Aguilella, V.M.; Verdiá-Báguena, C.; Alcaraz, A. Lipid Charge Regulation of Non-Specific Biological Ion Channels. Phys. Chem. Chem. Phys. 2014, 16, 3881–3893. [Google Scholar] [CrossRef] [PubMed]
- Cabrera-Garcia, D.; Bekdash, R.; Abbott, G.W.; Yazawa, M.; Harrison, N.L. The Envelope Protein of SARS-CoV-2 Increases Intra-Golgi PH and Forms a Cation Channel That Is Regulated by PH. J. Physiol. 2021, 599, 2851–2868. [Google Scholar] [CrossRef]
- Breitinger, U.; Ali, N.K.M.; Sticht, H.; Breitinger, H.G. Inhibition of SARS CoV Envelope Protein by Flavonoids and Classical Viroporin Inhibitors. Front. Microbiol. 2021, 12, 692426. [Google Scholar] [CrossRef] [PubMed]
- Xia, B.; Shen, X.; He, Y.; Pan, X.; Liu, F.L.; Wang, Y.; Yang, F.; Fang, S.; Wu, Y.; Duan, Z.; et al. SARS-CoV-2 Envelope Protein Causes Acute Respiratory Distress Syndrome (ARDS)-like Pathological Damages and Constitutes an Antiviral Target. Cell Res. 2021, 31, 847–860. [Google Scholar] [CrossRef]
- Alcaraz, A.; Nestorovich, E.M.; López, M.L.; García-Giménez, E.; Bezrukov, S.M.; Aguilella, V.M. Diffusion, Exclusion, and Specific Binding in a Large Channel: A Study of OmpF Selectivity Inversion. Biophys. J. 2009, 96, 56–66. [Google Scholar] [CrossRef] [Green Version]
- Antonides, L.H.; Hurst, Q.W.; Ives, C.M.; Ramberg, K.; Ostrovitsa, N.; Scanlan, E.; Caffrey, M.; Pitt, S.J.; Zachariae, U.; Kiefer, R. The SARS-CoV-2 Envelope (E) Protein Forms a Calcium- and Voltage-Activated Calcium Channel. bioRxiv 2022. [Google Scholar] [CrossRef]
- Ito, M.; Yanagi, Y.; Ichinohe, T. Encephalomyocarditis Virus Viroporin 2B Activates NLRP3 Inflammasome. PLoS Pathog. 2012, 8, e1002857. [Google Scholar] [CrossRef] [Green Version]
- Appenzeller-Herzog, C.; Hauri, H.P. The ER-Golgi Intermediate Compartment (ERGIC): In Search of Its Identity and Function. J. Cell Sci. 2006, 119, 2173–2183. [Google Scholar] [CrossRef] [Green Version]
- Murakami, T.; Ockinger, J.; Yu, J.; Byles, V.; McColl, A.; Hofer, A.M.; Horng, T. Critical Role for Calcium Mobilization in Activation of the NLRP3 Inflammasome. Proc. Natl. Acad. Sci. USA 2012, 109, 11282–11287. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Frey, T.K.; Yang, J.J. Viral Calciomics: Interplays between Ca2+ and Virus. Cell Calcium 2009, 46, 1–17. [Google Scholar] [CrossRef]
- Gillespie, D.; Boda, D. The Anomalous Mole Fraction Effect in Calcium Channels: A Measure of Preferential Selectivity. Biophys. J. 2008, 95, 2658–2672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillespie, D.; Boda, D.; He, Y.; Apel, P.; Siwy, Z.S. Synthetic Nanopores as a Test Case for Ion Channel Theories: The Anomalous Mole Fraction Effect without Single Filing. Biophys. J. 2008, 95, 609–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Queralt-Martín, M.; López, M.L.; Aguilella-Arzo, M.; Aguilella, V.M.; Alcaraz, A. Scaling Behavior of Ionic Transport in Membrane Nanochannels. Nano Lett. 2018, 18, 6604–6610. [Google Scholar] [CrossRef]
- Hille, B. Ion Channels of Excitable Membranes, 3rd ed.; Sinauer Associates, Oxford University Press: Sunderland, MA, USA, 2001; ISBN 978-0878933211. [Google Scholar]
- Largo, E.; Verdiá-Báguena, C.; Aguilella, V.M.; Nieva, J.L.; Alcaraz, A. Ion Channel Activity of the CSFV P7 Viroporin in Surrogates of the ER Lipid Bilayer. Biochim. Biophys. Acta Biomembr. 2016, 1858, 30–37. [Google Scholar] [CrossRef] [Green Version]
- Largo, E.; Queralt-Martín, M.; Carravilla, P.; Nieva, J.L.; Alcaraz, A. Single-Molecule Conformational Dynamics of Viroporin Ion Channels Regulated by Lipid-Protein Interactions. Bioelectrochemistry 2021, 137, 107641. [Google Scholar] [CrossRef]
- Leviatan, S.; Sawada, K.; Moriyama, Y.; Nelson, N. Combinatorial Method for Overexpression of Membrane Proteins in Escherichia Coli. J. Biol. Chem. 2010, 285, 23548–23556. [Google Scholar] [CrossRef] [Green Version]
- Studier, F.W. Stable Expression Clones and Auto-Induction for Protein Production in E. Coli. Methods Mol. Biol. 2014, 1091, 17–32. [Google Scholar] [CrossRef]
- Brown, P.H.; Schuck, P. Macromolecular Size-and-Shape Distributions by Sedimentation Velocity Analytical Ultracentrifugation. Biophys. J. 2006, 90, 4651–4661. [Google Scholar] [CrossRef] [Green Version]
- Brautigam, C.A. Calculations and Publication-Quality Illustrations for Analytical Ultracentrifugation Data. Methods Enzymol. 2015, 562, 109–133. [Google Scholar] [CrossRef] [PubMed]
- Surya, W.; Liu, Y.; Torres, J. The Cytoplasmic N-Terminal Tail of Zika Virus NS4A Protein Forms Oligomers in the Absence of Detergent or Lipids. Sci. Rep. 2023, 13, 7360. [Google Scholar] [CrossRef] [PubMed]
- Laue, T.M.; Shah, B.D.; Ridgeway, T.M.; Pelletier, S.L. Computer-Aided Interpretation of Analytical Sedimentation Data for Proteins. In Analytical Ultracentrifugation in Biochemistry and Polymer Science; Harding, S.E., Rowe, A.J., Horton, J.C., Eds.; Royal Society of Chemistry: Cambridge, UK, 1992; pp. 90–125. [Google Scholar]
- Burgess, N.K.; Stanley, A.M.; Fleming, K.G. Determination of Membrane Protein Molecular Weights and Association Equilibrium Constants Using Sedimentation Equilibrium and Sedimentation Velocity. Methods Cell Biol. 2008, 84, 181–211. [Google Scholar] [PubMed]
- Bezrukov, S.M.; Vodyanoy, I. Probing Alamethicin Channels with Water-Soluble Polymers. Effect on Conductance of Channel States. Biophys. J. 1993, 64, 16–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montal, M.; Mueller, P. Formation of Bimolecular Membranes from Lipid Monolayers and a Study of Their Electrical Properties. Proc. Natl. Acad. Sci. USA 1972, 69, 3561–3566. [Google Scholar] [CrossRef] [PubMed]

| Protein-Specific Parameters | |||
|---|---|---|---|
| E-TM | 6H-E-TR | E-FL | |
| Monomeric MW (Da) | 3490 | 9081 | 8542 |
| ῡ (20 °C) (mL/g) | 0.7929 | 0.7549 | 0.7656 |
| Frictional ratio, f/f0 | 1.8–3.0 | 1.5–2.7 | 1.5–2.5 |
| Predicted S-Value Range | |||
| E-TM | 6H-E-TR | E-FL | |
| Monomer (S) | 0.16–0.27 (0.21) | 0.42–0.76 (0.56) | 0.41–0.69 (0.55) |
| Dimer (S) | 0.25–0.43 (0.34) | 0.66–1.22 (0.93) | 0.65–1.11 (0.87) |
| Trimer (S) | 0.33–0.57 (0.45) | 0.87–1.60 (1.22) | 0.85–1.45 (1.14) |
| Tetramer (S) | 0.40–0.69 (0.54) | 1.05–1.93 (1.48) | 1.03–1.76 (1.38) |
| Pentamer (S) | 0.47–0.80 (0.63) | 1.22–2.24 (1.72) | 1.19–2.04 (1.60) |
| Decamer (S) | 0.74–1.27 (1.00) | 1.94–3.56 (2.73) | 1.90–3.23 (2.55) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Surya, W.; Tavares-Neto, E.; Sanchis, A.; Queralt-Martín, M.; Alcaraz, A.; Torres, J.; Aguilella, V.M. The Complex Proteolipidic Behavior of the SARS-CoV-2 Envelope Protein Channel: Weak Selectivity and Heterogeneous Oligomerization. Int. J. Mol. Sci. 2023, 24, 12454. https://doi.org/10.3390/ijms241512454
Surya W, Tavares-Neto E, Sanchis A, Queralt-Martín M, Alcaraz A, Torres J, Aguilella VM. The Complex Proteolipidic Behavior of the SARS-CoV-2 Envelope Protein Channel: Weak Selectivity and Heterogeneous Oligomerization. International Journal of Molecular Sciences. 2023; 24(15):12454. https://doi.org/10.3390/ijms241512454
Chicago/Turabian StyleSurya, Wahyu, Ernesto Tavares-Neto, Andrea Sanchis, María Queralt-Martín, Antonio Alcaraz, Jaume Torres, and Vicente M. Aguilella. 2023. "The Complex Proteolipidic Behavior of the SARS-CoV-2 Envelope Protein Channel: Weak Selectivity and Heterogeneous Oligomerization" International Journal of Molecular Sciences 24, no. 15: 12454. https://doi.org/10.3390/ijms241512454
APA StyleSurya, W., Tavares-Neto, E., Sanchis, A., Queralt-Martín, M., Alcaraz, A., Torres, J., & Aguilella, V. M. (2023). The Complex Proteolipidic Behavior of the SARS-CoV-2 Envelope Protein Channel: Weak Selectivity and Heterogeneous Oligomerization. International Journal of Molecular Sciences, 24(15), 12454. https://doi.org/10.3390/ijms241512454







