A Split-Marker System for CRISPR-Cas9 Genome Editing in Methylotrophic Yeasts
Abstract
:1. Introduction
2. Results
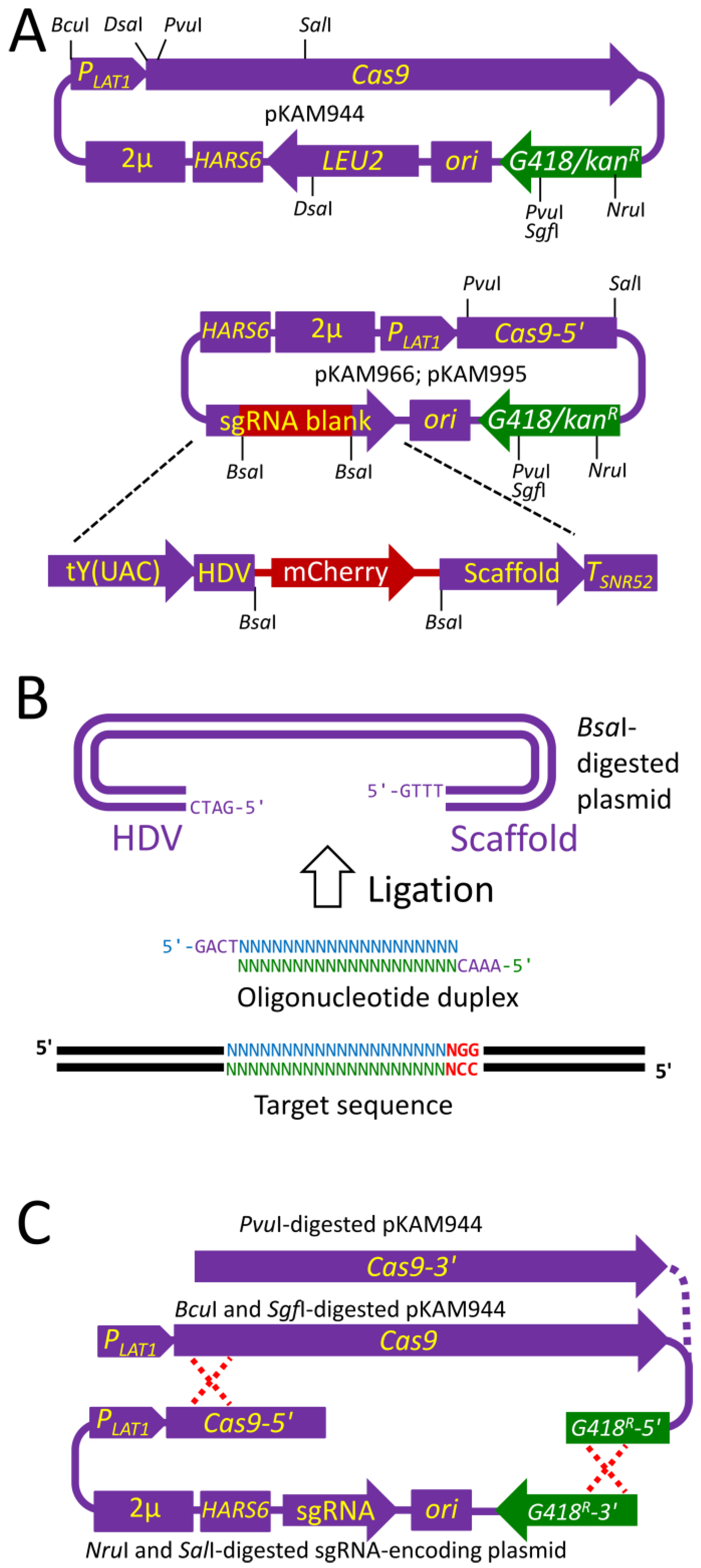
2.1. Design of the Plasmids Bearing the Components of the CRISPR-Cas9 Genome Editing System
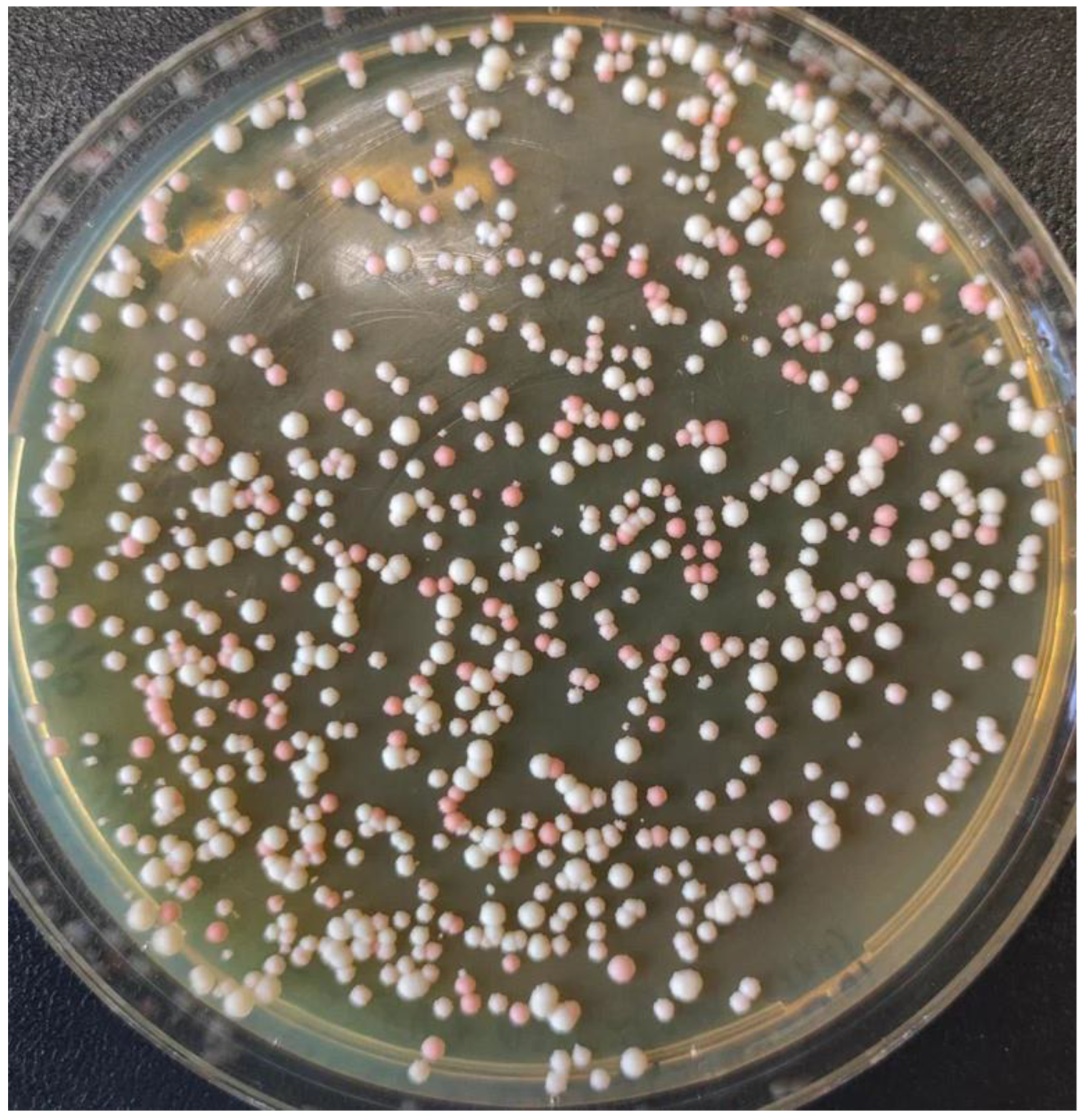
2.2. Inactivation of the MET8 Gene in O. polymorpha
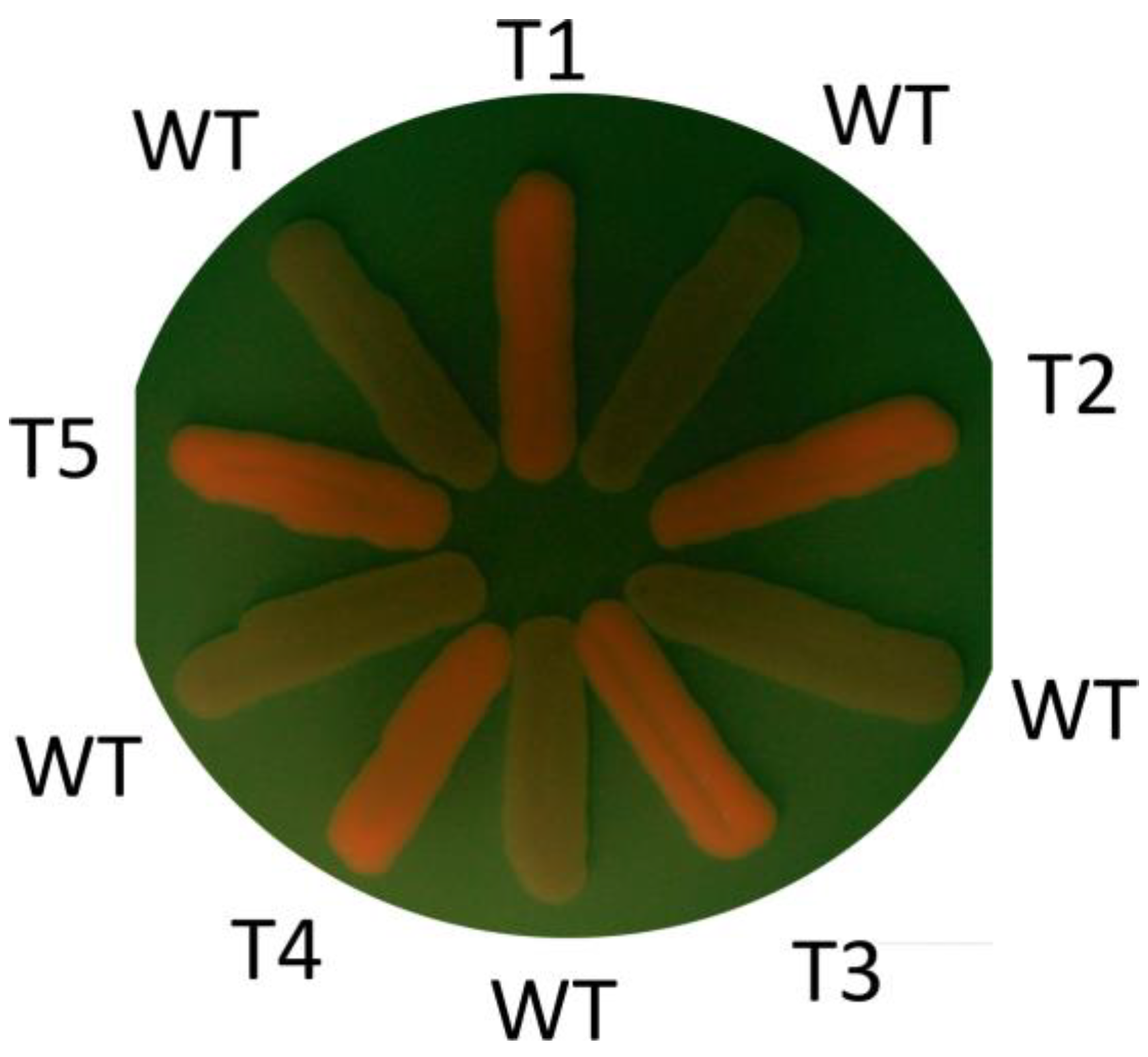
2.3. Inactivation of the ADE2 Gene in O. haglerorum
2.4. The Use of the LEU2 Selectable Marker to Transform O. polymorpha and O. parapolymorpha with sgRNA and Cas9-Encoding Plasmids
2.5. Inactivation of the MET8 Gene in K. phaffii
3. Discussion
4. Materials and Methods
4.1. Genetic Nomenclature Yeast Strains and Culture Conditions
4.2. Plasmids, Oligonucleotides and PCR Conditions
4.3. Yeast Transformation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wiedenheft, B.; Sternberg, S.H.; Doudna, J.A. RNA-Guided Genetic Silencing Systems in Bacteria and Archaea. Nature 2012, 482, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Bhaya, D.; Davison, M.; Barrangou, R. CRISPR-Cas Systems in Bacteria and Archaea: Versatile Small RNAs for Adaptive Defense and Regulation. Annu. Rev. Genet. 2011, 45, 273–297. [Google Scholar] [CrossRef] [PubMed]
- Terns, M.P.; Terns, R.M. CRISPR-Based Adaptive Immune Systems. Curr. Opin. Microbiol. 2011, 14, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Gasiunas, G.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas9–CrRNA Ribonucleoprotein Complex Mediates Specific DNA Cleavage for Adaptive Immunity in Bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, 2579–2584. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Aman Mohammadi, M.; Maximiano, M.R.; Hosseini, S.M.; Franco, O.L. CRISPR-Cas Engineering in Food Science and Sustainable Agriculture: Recent Advancements and Applications. Bioprocess Biosyst. Eng. 2023, 46, 483–497. [Google Scholar] [CrossRef]
- Kalds, P.; Zhou, S.; Cai, B.; Liu, J.; Wang, Y.; Petersen, B.; Sonstegard, T.; Wang, X.; Chen, Y. Sheep and Goat Genome Engineering: From Random Transgenesis to the CRISPR Era. Front. Genet. 2019, 10, 750. [Google Scholar] [CrossRef]
- Whitworth, K.M.; Lee, K.; Benne, J.A.; Beaton, B.P.; Spate, L.D.; Murphy, S.L.; Samuel, M.S.; Mao, J.; O’Gorman, C.; Walters, E.M.; et al. Use of the CRISPR/Cas9 System to Produce Genetically Engineered Pigs from In Vitro-Derived Oocytes and Embryos1. Biol. Reprod. 2014, 91, 78. [Google Scholar] [CrossRef]
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.-L. Simultaneous Editing of Three Homoeoalleles in Hexaploid Bread Wheat Confers Heritable Resistance to Powdery Mildew. Nat. Biotechnol. 2014, 32, 947–951. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, P.; Niu, Y.; Zhang, Y.; Wen, G.; Zhao, C.; Jiang, M. Evolution of the WRKY66 Gene Family and Its Mutations Generated by the CRISPR/Cas9 System Increase the Sensitivity to Salt Stress in Arabidopsis. Int. J. Mol. Sci. 2023, 24, 3071. [Google Scholar] [CrossRef]
- Wu, Y.; Zeng, J.; Roscoe, B.P.; Liu, P.; Yao, Q.; Lazzarotto, C.R.; Clement, K.; Cole, M.A.; Luk, K.; Baricordi, C.; et al. Highly Efficient Therapeutic Gene Editing of Human Hematopoietic Stem Cells. Nat. Med. 2019, 25, 776–783. [Google Scholar] [CrossRef]
- Gillmore, J.D.; Gane, E.; Taubel, J.; Kao, J.; Fontana, M.; Maitland, M.L.; Seitzer, J.; O’Connell, D.; Walsh, K.R.; Wood, K.; et al. CRISPR-Cas9 In Vivo Gene Editing for Transthyretin Amyloidosis. N. Engl. J. Med. 2021, 385, 493–502. [Google Scholar] [CrossRef]
- Li, T.; Yang, Y.; Qi, H.; Cui, W.; Zhang, L.; Fu, X.; He, X.; Liu, M.; Li, P.; Yu, T. CRISPR/Cas9 Therapeutics: Progress and Prospects. Signal Transduct. Target. Ther. 2023, 8, 36. [Google Scholar] [CrossRef]
- Cai, G.; Lin, Z.; Shi, S. Development and Expansion of the CRISPR/Cas9 Toolboxes for Powerful Genome Engineering in Yeast. Enzyme Microb. Technol. 2022, 159, 110056. [Google Scholar] [CrossRef]
- Gao, Y.; Zhao, Y. Self-Processing of Ribozyme-Flanked RNAs into Guide RNAs in Vitro and in Vivo for CRISPR-Mediated Genome Editing. J. Integr. Plant Biol. 2014, 56, 343–349. [Google Scholar] [CrossRef]
- Weninger, A.; Hatzl, A.M.; Schmid, C.; Vogl, T.; Glieder, A. Combinatorial Optimization of CRISPR/Cas9 Expression Enables Precision Genome Engineering in the Methylotrophic Yeast Pichia Pastoris. J. Biotechnol. 2016, 235, 139–149. [Google Scholar] [CrossRef]
- Ryan, O.W.; Skerker, J.M.; Maurer, M.J.; Li, X.; Tsai, J.C.; Poddar, S.; Lee, M.E.; DeLoache, W.; Dueber, J.E.; Arkin, A.P.; et al. Selection of Chromosomal DNA Libraries Using a Multiplex CRISPR System. Elife 2014, 3, e03703. [Google Scholar] [CrossRef]
- Leão, A.N.; Kiel, J.A.K.W. Peroxisome Homeostasis in Hansenula Polymorpha. FEMS Yeast Res. 2003, 4, 131–139. [Google Scholar] [CrossRef]
- Moon, H.; Cheon, S.A.; Kim, H.; Agaphonov, M.O.; Kwon, O.; Oh, D.; Kim, J.; Kang, H.A. Hansenula Polymorpha Hac1p Is Critical to Protein N -Glycosylation Activity Modulation, as Revealed by Functional and Transcriptomic Analyses. Appl. Environ. Microbiol. 2015, 81, 6982–6993. [Google Scholar] [CrossRef]
- Siverio, J.M. Assimilation of Nitrate by Yeasts. FEMS Microbiol. Rev. 2002, 26, 277–284. [Google Scholar] [CrossRef]
- Viigand, K.; Alamae, T. Further Study of the Hansenula Polymorpha MAL Locus: Characterization of the alpha-Glucoside Permease Encoded by the HpMAL2 Gene. FEMS Yeast Res. 2007, 7, 1134–1144. [Google Scholar] [CrossRef] [PubMed]
- Seike, T.; Narazaki, Y.; Kaneko, Y.; Shimizu, H.; Matsuda, F. Random Transfer of Ogataea Polymorpha Genes into Saccharomyces Cerevisiae Reveals a Complex Background of Heat Tolerance. J. Fungi 2021, 7, 302. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, H.; Neuner, A.; Rüthnick, D.; Schiebel, E.; Pereira, G.; Kaneko, Y. Polo-like Kinase Cdc5 Regulates Spc72 Recruitment to Spindle Pole Body in the Methylotrophic Yeast Ogataea Polymorpha. Elife 2017, 6, e24340. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Yu, W.; Zhai, X.; Yao, L.; Guo, X.; Gao, J.; Zhou, Y.J. Characterizing and Engineering Promoters for Metabolic Engineering of Ogataea Polymorpha. Synth. Syst. Biotechnol. 2022, 7, 498–505. [Google Scholar] [CrossRef]
- Kurylenko, O.O.; Ruchala, J.; Hryniv, O.B.; Abbas, C.A.; Dmytruk, K.V.; Sibirny, A.A. Metabolic Engineering and Classical Selection of the Methylotrophic Thermotolerant Yeast Hansenula Polymorpha for Improvement of High-Temperature Xylose Alcoholic Fermentation. Microb. Cell Fact. 2014, 13, 122. [Google Scholar] [CrossRef]
- Dmytruk, K.V.; Sibirny, A.A. Metabolic Engineering of the Yeast Hansenula Polymorpha for the Construction of Efficient Ethanol Producers. Cytol. Genet. 2013, 47, 329–342. [Google Scholar] [CrossRef]
- Numamoto, M.; Maekawa, H.; Kaneko, Y. Efficient Genome Editing by CRISPR/Cas9 with a TRNA-SgRNA Fusion in the Methylotrophic Yeast Ogataea Polymorpha. J. Biosci. Bioeng. 2017, 124, 487–492. [Google Scholar] [CrossRef]
- Gao, J.; Gao, N.; Zhai, X.; Zhou, Y.J. Recombination Machinery Engineering for Precise Genome Editing in Methylotrophic Yeast Ogataea Polymorpha. iScience 2021, 24, 102168. [Google Scholar] [CrossRef]
- Wang, L.; Deng, A.; Zhang, Y.; Liu, S.; Liang, Y.; Bai, H.; Cui, D.; Qiu, Q.; Shang, X.; Yang, Z.; et al. Efficient CRISPR–Cas9 Mediated Multiplex Genome Editing in Yeasts. Biotechnol. Biofuels 2018, 11, 277. [Google Scholar] [CrossRef]
- Engler, C.; Kandzia, R.; Marillonnet, S. A One Pot, One Step, Precision Cloning Method with High Throughput Capability. PLoS ONE 2008, 3, e3647. [Google Scholar] [CrossRef]
- Karginov, A.V.; Alexandrov, A.I.; Kushnirov, V.V.; Agaphonov, M.O. Perturbations in the Heme and Siroheme Biosynthesis Pathways Causing Accumulation of Fluorescent Free Base Porphyrins and Auxotrophy in Ogataea Yeasts. J. Fungi 2021, 7, 884. [Google Scholar] [CrossRef]
- Shaw, W. Quick and Easy CRISPR Engineering in Saccharomyces Cerevisiae. Available online: https://benchling.com/pub/ellis-crispr-tools (accessed on 2 May 2023).
- Kim, M.W.; Kim, E.J.; Kim, J.; Park, J.-S.; Oh, D.-B.; Shimma, Y.; Chiba, Y.; Jigami, Y.; Rhee, S.K.; Kang, H.A. Functional Characterization of the Hansenula Polymorpha HOC1, OCH1, and OCR1 Genes as Members of the Yeast OCH1 Mannosyltransferase Family Involved in Protein Glycosylation. J. Biol. Chem. 2006, 281, 6261–6272. [Google Scholar] [CrossRef]
- Kim, H.; Thak, E.J.; Lee, D.J.; Agaphonov, M.O.; Kang, H.A. Hansenula Polymorpha Pmt4p Plays Critical Roles in O-Mannosylation of Surface Membrane Proteins and Participates in Heteromeric Complex Formation. PLoS ONE 2015, 10, e0129914. [Google Scholar] [CrossRef]
- Agaphonov, M.O.; Sokolov, S.S.; Romanova, N.V.; Sohn, J.H.; Kim, S.Y.; Kalebina, T.S.; Choi, E.S.; Ter-Avanesyan, M.D. Mutation of the Protein-O-Mannosyltransferase Enhances Secretion of the Human Urokinase-Type Plasminogen Activator in Hansenula Polymorpha. Yeast 2005, 22, 1037–1047. [Google Scholar] [CrossRef]
- Agaphonov, M.O.; Packeiser, A.N.; Chechenova, M.B.; Choi, E.S.; Ter-Avanesyan, M.D. Mutation of the Homologue of GDP-Mannose Pyrophosphorylase Alters Cell Wall Structure, Protein Glycosylation and Secretion in Hansenula Polymorpha. Yeast 2001, 18, 391–402. [Google Scholar] [CrossRef]
- Bogdanova, A.I.; Kustikova, O.S.; Agaphonov, M.O.; Ter-Avanesyan, M.D. Sequences of Saccharomyces Cerevisiae 2 μm DNA Improving Plasmid Partitioning In Hansenula Polymorpha. Yeast 1998, 14, 1–9. [Google Scholar] [CrossRef]
- Bunting, M.; Bernstein, K.E.; Greer, J.M.; Capecchi, M.R.; Thomas, K.R. Targeting Genes for Self-Excision in the Germ Line. Genes Dev. 1999, 13, 1524–1528. [Google Scholar] [CrossRef]
- Kaczmarczyk, S.J.; Green, J.E. A Single Vector Containing Modified Cre Recombinase and LOX Recombination Sequences for Inducible Tissue-Specific Amplification of Gene Expression. Nucleic Acids Res. 2001, 29, E56. [Google Scholar] [CrossRef]
- Agaphonov, M.; Alexandrov, A. Self-Excising Integrative Yeast Plasmid Vectors Containing an Intronated Recombinase Gene. FEMS Yeast Res. 2014, 14, 1048–1054. [Google Scholar] [CrossRef]
- Agaphonov, M.O. Improvement of a Yeast Self-Excising Integrative Vector by Prevention of Expression Leakage of the Intronated Cre Recombinase Gene during Plasmid Maintenance in Escherichia Coli. FEMS Microbiol. Lett. 2017, 364, 22–25. [Google Scholar] [CrossRef]
- Gellissen, G. (Ed.) Hansenula Polymorpha: Biology and Applications; Wiley: Weinheim, Germany, 2002; ISBN 9783527303410. [Google Scholar]
- Sohn, J.H.; Choi, E.S.; Kim, C.H.; Agaphonov, M.O.; Ter-Avanesyan, M.D.; Rhee, J.S.; Rhee, S.K. A Novel Autonomously Replicating Sequence (ARS) for Multiple Integration in the Yeast Hansenula Polymorpha DL-1. J. Bacteriol. 1996, 178, 4420–4428. [Google Scholar] [CrossRef] [PubMed]
- Naumov, G.I.; Naumova, E.S.; Lee, C.-F. Ogataea Haglerorum Sp. Nov., a Novel Member of the Species Complex, Ogataea (Hansenula) Polymorpha. Int. J. Syst. Evol. Microbiol. 2017, 67, 2465–2469. [Google Scholar] [CrossRef] [PubMed]
- Agaphonov, M.; Romanova, N.; Choi, E.-S.; Ter-Avanesyan, M. A Novel Kanamycin/G418 Resistance Marker for Direct Selection of Transformants in Escherichia Coli and Different Yeast Species. Yeast 2010, 27, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Gassler, T.; Heistinger, L.; Mattanovich, D.; Gasser, B.; Prielhofer, R. CRISPR/Cas9-Mediated Homology-Directed Genome Editing in Pichia pastoris. In Recombinant Protein Production in Yeast. Methods in Molecular Biology; Gasser, B., Mattanovich, D., Eds.; Humana Press: New York, NY, USA, 2019; pp. 211–225. ISBN 978-1-4939-9024-5. [Google Scholar]
- Gietz, R.D.; Schiestl, R.H. Quick and Easy Yeast Transformation Using the LiAc/SS Carrier DNA/PEG Method. Nat. Protoc. 2007, 2, 35–37. [Google Scholar] [CrossRef]
- Saraya, R.; Gidijala, L.; Veenhuis, M.; van der Klei, I.J. Tools for Genetic Engineering of the Yeast Hansenula polymorpha. In Yeast Metabolic Engineering: Methods and Protocols; Mapelli, V., Ed.; Humana: New York, NY, USA, 2014; pp. 43–62. ISBN 9781493905621. [Google Scholar]
- Lin-Cereghino, J.; Naranjo, C.A.; Lin-Cereghino, G.P. Competent Cell Preparation and Transformation of Pichia pastoris. In Yeast Metabolic Engineering: Methods and Protocols; Mapelli, V., Bettiga, M., Eds.; Springer: New York, NY, USA, 2022; pp. 113–120. [Google Scholar]


| Name | 5′-3′ Sequence | Purpose |
|---|---|---|
| oriA | CCTATGGAAAAACGCCAGCAA | PCR for plasmid construction |
| ScLEU2_989HpaI | TTTGTTAACTGCATCTTCAATGGC | PCR for plasmid construction |
| OpoMET8crU | GACTAGCAGATGCACAGATCACAG | OpoMET8 sgRNA |
| OpoMET8crL | AAACCTGTGATCTGTGCATCTGCT | OpoMET8 sgRNA |
| MET8opoAU1 | GTCCTTGGAGACACCTTACC | OpoMET8 PCR analysis |
| MET8opoAL1 | CCGCTCGAAATGCGCTCTAT | OpoMET8 PCR analysis |
| KpMET8F | GATCTTGGGTCAGTAGTAAGACAG | KpMET8 sgRNA |
| KpMET8R | AAACCTGTCTTACTACTGACCCAA | KpMET8 sgRNA |
| OhADE2F1 | GACTTGGAGACGGATAGATCCGGA | OhADE2 sgRNA |
| OhADE2R1 | AAACTCCGGATCTATCCGTCTCCA | OhADE2 sgRNA |
| OhADE2F2 | GACTAGCAGTGCTCTCAACAGCGG | OhADE2 sgRNA |
| OhADE2R2 | AAACCCGCTGTTGAGAGCACTGCT | OhADE2 sgRNA |
| Up-ADE2-F | GATGTCGAAGTCAACAAAATCCAGG | Construction of donor DNA for OhADE2 deletion |
| Up-ADE2-R | CCTACGACCTTCGAGTCCATGATAC | Construction of donor DNA for OhADE2 deletion |
| Dn-ADE2-F | TACATTAATTTAATTAGTATCATGGACTCGAAGGTCGTAGGGGCTCTGTTGGCTACGAGGAG | Construction of donor DNA for OhADE2 deletion |
| Dn-ADE2-R | CTCCATTTCCACCCTTTCCCGAC | Construction of donor DNA for OhADE2 deletion |
| Up-ADE2-sq-F | CCTTCTGTCGTCTACCGTTCTCTGTC | OhADE2 PCR and sequencing analyses |
| Dn-ADE2-sq-R | CGACAGCGGACACATAGACGTTGC | OhADE2 PCR and sequencing analyses |
| ADE2-4-F | GTATCATGGACTCGAAGGTCGTAGG | OhADE2 PCR and sequencing analyses |
| ADE2-847-R | CTCAAACTGGCTCGTAACACACGC | OhADE2 PCR and sequencing analyses |
| PMT1_786U | GACTTCCCACAACCATCTGTACGA | OpaPMT1 sgRNA |
| PMT1_786L | AAACTCGTACAGATGGTTGTGGGA | OpaPMT1 sgRNA |
| Experiment | pKAM944 Cut | Donor DNA | Preincubation (h) | # of Transformants | # of Met− | % of Met− |
|---|---|---|---|---|---|---|
| 1 | PvuI | + | 2 | 110 | 22 | 20 |
| 1 | PvuI | - | 2 | 49 | 0 | 0 |
| 1 | SgfI BcuI | + | 2 | 244 | 29 | 12 |
| 1 | SgfI BcuI | - | 2 | 158 | 13 | 8 |
| 2 | SgfI BcuI | + | 1 | 38 | 0 | 0 |
| 2 | SgfI BcuI | + | 2 | 49 | 3 | 6 |
| 2 | SgfI BcuI | + | 3 | 43 | 6 | 14 |
| 2 | SgfI BcuI | - | 3 | 88 | 16 | 18 |
| 2 | PvuI | - | 3 | 81 | 11 | 14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karginov, A.V.; Tarutina, M.G.; Lapteva, A.R.; Pakhomova, M.D.; Galliamov, A.A.; Filkin, S.Y.; Fedorov, A.N.; Agaphonov, M.O. A Split-Marker System for CRISPR-Cas9 Genome Editing in Methylotrophic Yeasts. Int. J. Mol. Sci. 2023, 24, 8173. https://doi.org/10.3390/ijms24098173
Karginov AV, Tarutina MG, Lapteva AR, Pakhomova MD, Galliamov AA, Filkin SY, Fedorov AN, Agaphonov MO. A Split-Marker System for CRISPR-Cas9 Genome Editing in Methylotrophic Yeasts. International Journal of Molecular Sciences. 2023; 24(9):8173. https://doi.org/10.3390/ijms24098173
Chicago/Turabian StyleKarginov, Azamat V., Marina G. Tarutina, Anastasia R. Lapteva, Maria D. Pakhomova, Artur A. Galliamov, Sergey Y. Filkin, Alexey N. Fedorov, and Michael O. Agaphonov. 2023. "A Split-Marker System for CRISPR-Cas9 Genome Editing in Methylotrophic Yeasts" International Journal of Molecular Sciences 24, no. 9: 8173. https://doi.org/10.3390/ijms24098173
APA StyleKarginov, A. V., Tarutina, M. G., Lapteva, A. R., Pakhomova, M. D., Galliamov, A. A., Filkin, S. Y., Fedorov, A. N., & Agaphonov, M. O. (2023). A Split-Marker System for CRISPR-Cas9 Genome Editing in Methylotrophic Yeasts. International Journal of Molecular Sciences, 24(9), 8173. https://doi.org/10.3390/ijms24098173






