Molecular Characterization of Nine TRAF Genes in Yellow Catfish (Pelteobagrus fulvidraco) and Their Expression Profiling in Response to Edwardsiella ictaluri Infection
Abstract
:1. Introduction
2. Results
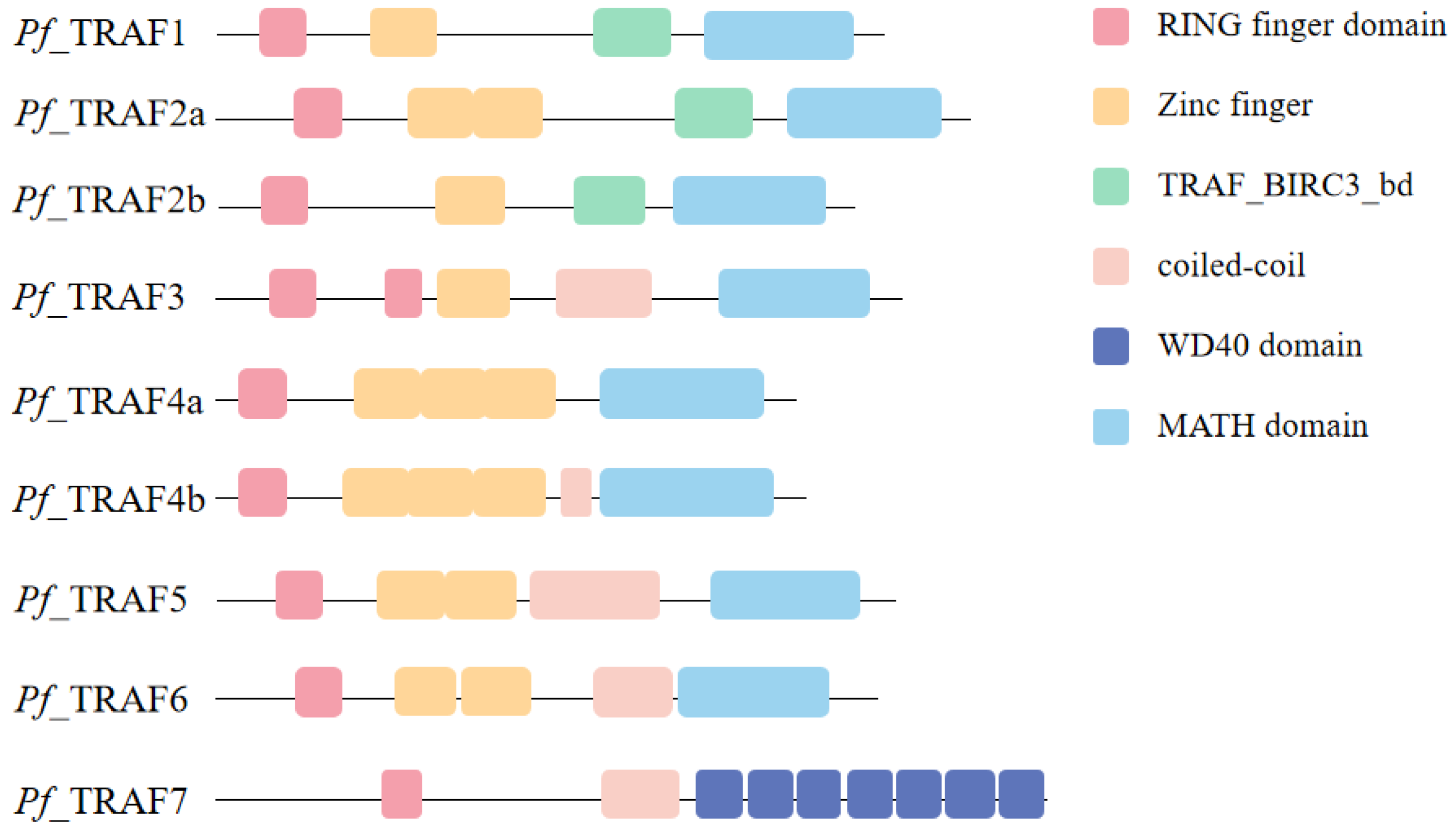
2.1. Protein Structural Characterization of Pf_TRAF1–7
2.2. Gene Structure and Motif Composition Analysis
2.3. Phylogenetic and Syntenic Analysis
2.4. Tissue Expression Profiling of Pf_TRAF Genes
2.5. The Involvement of Pf_TRAF Genes in Response to E. ictaluri Infection
2.6. Subcellular Localization
3. Discussion
4. Materials and Methods
4.1. Experimental Fish
4.2. Molecular Cloning and Bioinformatic Analysis
4.3. Tissue Collection in Healthy Fish and E. ictaluri Infected Fish
4.4. RNA Extraction, cDNA Synthesis and Quantitative Real-Time PCR Analysis
4.5. Plasmid Construction
4.6. Transfection of HEK293T Cells
4.7. Data Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chung, J.Y.; Park, Y.C.; Ye, H.; Wu, H. TRAFs are not created equal: Common and distinct molecular mechanisms of TRAF-mediated signal transduction. J. Cell. Sci. 2002, 115 Pt 4, 679–688. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Jiang, Y.; Wang, A.; Husain, M.; Xu, Q.Q.; Secombes, C.J. Identification of the salmonid IL-17A/F1a/b, IL-17A/F2b, IL-17A/F3 and IL-17N genes and analysis of their expression following in vitro stimulation and infection. Immunogenetics 2015, 67, 395–412. [Google Scholar] [CrossRef] [PubMed]
- Inoue, J.; Ishida, T.; Tsukamoto, N.; Kobayashi, N.; Naito, A.; Azuma, S.; Yamamoto, T. Tumor necrosis factor receptor-associated factor (TRAF) family: Adapter proteins that mediate cytokine signaling. Exp. Cell Res. 2000, 254, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Wajant, H.; Henkler, F.; Scheurich, P. The TNF-receptor-associated factor family: Scaffold molecules for cytokine receptors, kinases and their regulators. Cell. Signal. 2001, 13, 389–400. [Google Scholar] [CrossRef]
- Xu, Y.C.; Wu, R.F.; Gu, Y.; Yang, Y.S.; Yang, M.C.; Nwariaku, F.E.; Terada, L.S. Involvement of traf4 in oxidative activation of C-jun N-terminal kinase. J. Biol. Chem. 2002, 277, 28051–28057. [Google Scholar] [CrossRef]
- Rothe, M.; Wong, S.C.; Henzel, W.J.; Goeddel, D.V. A novel family of putative signal transducers associated with the cytoplasmic domain of the 75 kDa tumor necrosis factor receptor. Cell 1994, 78, 681–692. [Google Scholar] [CrossRef]
- Wajant, H.; Mühlenbeck, F.; Scheurich, P. Identification of a TRAF (TNF receptor-associated factor) gene in Caenorhabditis elegans. J. Mol. Evol. 1998, 47, 656–662. [Google Scholar] [CrossRef]
- Xu, L.G.; Li, L.Y.; Shu, H.B. TRAF7 potentiates MEKK3-induced AP1 and CHOP activation and induces apoptosis. J. Biol. Chem. 2004, 279, 17278–17282. [Google Scholar] [CrossRef]
- Takeuchi, M.; Rothe, M.; Goeddel, D.V. Anatomy of TRAF2. J. Biol. Chem. 1996, 271, 19935–19942. [Google Scholar] [CrossRef]
- Rothe, M.; Sarma, V.; Dixit, V.M.; Goeddel, D.V. TRAF2-mediated activation of NF-kappa B by TNF receptor 2 and CD40. Science 1995, 269, 1424–1427. [Google Scholar] [CrossRef]
- Zapata, J.M.; Martínez-García, V.; Lefebvre, S. Phylogeny of the TRAF/MATH domain. Adv. Exp. Med. Biol. 2007, 597, 1–24. [Google Scholar] [PubMed]
- Ha, H.; Han, D.; Choi, Y. TRAF-mediated TNFR-family signaling. Curr. Protoc. Immunol. 2009, 87, 11.9D.1–11.9D.19. [Google Scholar] [CrossRef]
- Zotti, T.; Vito, P.; Stilo, R. The seventh ring: Exploring TRAF7 functions. J. Cell Physiol. 2012, 227, 1280–1284. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Li, J.; Ling, G. Research progress of tumor necrosis factor receptor-related factors. China Med. Herald. 2017, 14, 11. [Google Scholar]
- Mackay, J.P.; Crossley, M. Zinc fingers are sticking together. Trends Biochem. Sci. 1998, 23, 1–4. [Google Scholar] [CrossRef]
- Locksley, R.M.; Killeen, N.; Lenardo, M.J. The TNF and TNF receptor superfamilies: Integrating mammalian biology. Cell 2001, 104, 487–501. [Google Scholar] [CrossRef]
- Arch, R.H.; Gedrich, R.W.; Thompson, C.B. Tumor necrosis factor receptor-associated factors (TRAFs)-a family of adapter proteins that regulates life and death. Genes Dev. 1998, 12, 2821–2830. [Google Scholar] [CrossRef]
- Hoebe, K.; Beutler, B. TRAF3: A new component of the TLR-signaling apparatus. Trends. Mol. Med. 2006, 12, 187–189. [Google Scholar] [CrossRef]
- Sato, S.; Sugiyama, M.; Yamamoto, M.; Watanabe, Y.; Kawai, T.; Takeda, K.; Akira, S. Toll/IL-1 receptor domain-containing adaptor inducing IFN-beta (TRIF) associates with TNF receptor-associated factor 6 and TANK-Binding kinase 1, and activates two distinct transcription factors, NF-kappa Band IFN-regulatory factor-3, in the toll-like receptor signaling. J. Immunol. 2003, 171, 4304–4310. [Google Scholar]
- Takeshita, F.; Ishii, K.J.; Kobiyama, K.; Kojima, Y.; Coban, C.; Sasaki, S.; Ishii, N.; Klinman, D.M.; Okuda, K.; Akira, S.; et al. TRAF4 acts as a silencer in TLR-mediated signaling through the association with TRAF6 and TRIF. Eur. J. Immunol. 2005, 35, 2477–2485. [Google Scholar] [CrossRef]
- McCarthy, J.V.; Ni, J.; Dixit, V.M. RIP2 is a novel NF-kappaB-activating and cell death-inducing kinase. J. Biol. Chem. 1998, 273, 16968–16975. [Google Scholar] [CrossRef] [PubMed]
- Guan, K.; Wei, C.; Zheng, Z.; Song, T.; Wu, F.X.; Zhang, Y.H.; Cao, Y.; Ma, S.L.; Chen, W.; Xu, Q.B.; et al. MAVS promotes inflammasome activation by targeting ASC for K63-linked ubiquitination via the E3 ligase TRAF3. J. Immunol. 2015, 194, 4880–4890. [Google Scholar] [CrossRef] [PubMed]
- Marinis, J.M.; Hutti, J.E.; Homer, C.R.; Cobb, B.A.; Cantley, L.C.; McDonald, C.; Abbott, D.W. IκB kinaseα phosphorylation of TRAF4 downregulates innate immune signaling. Mol. Cell Biol. 2012, 32, 2479–2489. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, R.; Takaesu, G.; Yoshida, H.; Okamoto, F.; Yoshioka, T.; Choi, Y.; Akira, S.; Kawai, T.; Yoshimura, A.; Kobayashi, T. TRAF6 and MEKK1 play a pivotal role in the RIG-I-like helicase antiviral pathway. J. Biol. Chem. 2008, 283, 36211–36220. [Google Scholar] [CrossRef]
- Dhillon, B.; Aleithan, F.; Abdul-Sater, Z.; Abdul-Sater, A.A. The evolving role of TRAFs in mediating inflammatory responses. Front. Immunol. 2019, 10, 104. [Google Scholar] [CrossRef]
- Fang, R.; Jiang, Q.F.; Zhou, X.; Wang, C.G.; Guan, Y.K.; Tao, J.L.; Xi, J.Z.; Feng, J.M.; Jiang, Z.F. MAVS activates TBK1 and IKK epsilon through TRAFs in NEMO dependent and independent manner. PLoS Pathog. 2017, 13, e1006720. [Google Scholar] [CrossRef]
- Liu, S.Q.; Chen, J.Q.; Cai, X.; Wu, J.X.; Chen, X.; Wu, Y.T.; Sun, L.J.; Chen, Z.J. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. Elife 2013, 2, e00785. [Google Scholar] [CrossRef]
- Wu, B.; Hur, S. How RIG-I like receptors activate MAVS. Curr. Opin. Virol. 2015, 12, 91–98. [Google Scholar] [CrossRef]
- Li, K.M.; Li, M.; Wang, N.; Chen, Y.D.; Xu, X.W.; Xu, W.T.; Wang, L.; Chen, S.L. Genome-wide identification, characterization, and expression analysis of the TRAF gene family in Chinese tongue sole (Cynoglossus semilaevis). Fish Shellfish Immunol. 2020, 96, 13–25. [Google Scholar] [CrossRef]
- Han, M.; Liu, Y.X.; Jin, C.F.; Wang, X.G.; Song, W.H.; Zhang, Q.Q. Genome-wide identification, characterization and expression profiling of TRAF family genes in Sebastes schlegelii. Fish Shellfish. Immunol. 2022, 127, 203–210. [Google Scholar] [CrossRef]
- Han, X.Q.; Gao, F.Y.; Lu, M.X.; Liu, Z.G.; Cao, J.M.; Wang, M.; Yi, M.M.; Zhang, D.F. Cloning, expression and functional analysis of TRAF4 gene in Nile tilapia. J. Agric. Biol. 2019, 27, 381–392. [Google Scholar]
- Han, X.Q.; Gao, F.Y.; Liu, Z.G.; Liu, Z.G.; Cao, J.M.; Wang, M.; Yi, M.M.; Zhang, D.F. Expression profiles and signal transduction of TRAF6 in Nile Tilapia. Acta Hydrobiol. Sin. 2022, 46, 116–125. [Google Scholar]
- Xia, H.L.; Wang, Z.W.; Li, Y.; Chen, W.J.; Long, M.; Yu, D.P.; Cheng, J.; Xia, L.Q.; Lu, Y.S. Characterization of tumor necrosis factor receptor associated factor3 (TRAF3) in Nile Tilapia: Expression profiles and functions in NF-κB pathway. Acta Hydrobiol. Sin. 2023, 47, 308–315. [Google Scholar]
- Wu, S.P.; Ou, M.; Li, K.B.; Xu, Q.Q.; Xu, H.Y. Analysis of fbxo32 gene expression in the liver of TRAF6-null zebrafish. Acta Hydrobiol. Sin. 2021, 45, 945–950. [Google Scholar]
- Lu, R.H.; Chang, Z.G.; Sun, J.; Yang, F.; Nie, G.X.; Ji, H. Molecular cloning, expression and functional characterization of tumor necrosis factor (TNF) receptor-associated factor (TRAF)-interacting protein (TRIP) in grass carp, Ctenopharyngodon idella. Fish Shellfish Immunol. 2016, 57, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.Y.; Xiao, L.Y.; Kou, H.Y.; Yang, C.G.; Guo, P.H.; He, W.; Tian, D.Y.; Wu, K.; Cheng, Z.Q.; Song, X.H. Different routes of Aeromonas hydrophila infection lead to differential grass carp interleukin-17 family gene expression patterns during intestinal inflammation. Aquaculture 2020, 529, 735607. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Song, L.P. Research progress in the immune system of fish. HeBei Fish. 2018, 290, 49–56. [Google Scholar]
- Xie, P. TRAF molecules in cell signaling and in human diseases. J. Mol. Signal. 2013, 8, 7. [Google Scholar] [CrossRef]
- Wajant, H.; Scheurich, P. Tumor necrosis factor receptor-associated factor (TRAF) 2 and its role in TNF signaling. Int. J. Biochem. Cell Biol. 2001, 33, 19–32. [Google Scholar] [CrossRef]
- Arron, J.R.; Walsh, M.C.; Choi, Y.W. TRAF-mediated TNFR-family signaling. Curr. Protoc. Immunol. 2002, 51, 11–19. [Google Scholar] [CrossRef]
- Glasauer, S.M.K.; Neuhauss, S.C.F. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genomics. 2014, 289, 1045–1060. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Cheng, G.; Baltimore, D. Targeted disruption of TRAF3 leads to postnatal lethality and defective T-dependent immune responses. Immunity 1996, 5, 407–415. [Google Scholar] [CrossRef]
- Yeh, W.C.; Hakem, R.; Woo, M.; Mak, T.W. Gene targeting in the analysis of mammalian apoptosis and TNF receptor superfamily signaling. Immunol. Rev. 1999, 169, 283–302. [Google Scholar] [CrossRef]
- Wu, R.F.; Xu, Y.C.; Ma, Z.Y.; Nwariaku, F.E.; Sarosi, G.A., Jr.; Terada, L.S. Subcellular targeting of oxidants during endothelial cell migration. J. Cell Biol. 2005, 171, 893–904. [Google Scholar] [CrossRef]
- Wang, X.F.; Gao, S.; Hao, Z.X.; Tang, T.; Liu, F.S. Involvement of TRAF6 in regulating immune defense and ovarian development in Musca domestica. Int. J. Biol. Macromol. 2020, 15, 1262–1271. [Google Scholar] [CrossRef] [PubMed]
- Inoue, J.; Yagi, S.; Ishikawa, K.; Azuma, S.; Ikawa, S.; Semba, K. Identification and characterization of Xenopus laevis homologs of mammalian TRAF6 and its binding protein TIFA. Gene 2005, 358, 53–59. [Google Scholar] [CrossRef]
- Kalkan, T.; Iwasaki, Y.; Park, C.Y.; Thomsen, G.H. Tumor necrosis factor-receptor-associated factor-4 is a positive regulator of transforming growth factor-β signaling that affects neural crest formation. Mol. Biol. Cell 2009, 20, 3436–3450. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.Q.; Pang, Y.; Li, Q.W. Comprehensive evolutionary analysis of lamprey TNFR associated factors (TRAFs) and receptor-interacting protein kinase (RIPKs) and insights into the functional characterization of TRAF3/6 and RIPK1. Front. Immunol. 2020, 11, 663. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.L.; Feng, Z.Q.; Zeng, S.Q.; Li, S.M.; Zhu, Q.; Liu, Y.P. Molecular cloning and expression analysis of TRAF3 in chicken. Genet. Mol. Res. 2015, 14, 4408–4419. [Google Scholar] [CrossRef]
- Masson, R.; Regnier, C.H.; Chenard, M.P.; Wendling, C.; Mattei, M.G.; Tomasetto, C.; Rio, M.C. Tumor necrosis factor receptor associated factor 4 (TRAF4) expression pattern during mouse development. Mech. Dev. 1998, 71, 187–191. [Google Scholar] [CrossRef]
- Mizushima, S.I.; Fujita, M.K.; Ishida, T.; Azuma, S.; Kato, K.; Hirai, M.; Otsuka, M.; Yamamoto, T.; Inoue, J. Cloning and characterization of a cDNA encoding the human homolog of tumor necrosis factor receptor-associated factor 5 (TRAF5). Gene 1998, 207, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Salinas, I. The mucosal immune system of teleost fish. Biology 2015, 4, 525–539. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.W.; Huang, Y.; Li, Y.; Wang, B.; Lu, Y.S.; Xia, L.Q.; Tang, J.F.; Jian, J.C. Biological characterization, expression, and functional analysis of tumor necrosis factor receptor-associated factor 6 in Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 2018, 80, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Han, R.; Ni, L.Y.; Luo, X.C.; Li, A.X.; Dan, X.M.; Tsim, K.W.K.; Li, Y.W. Molecular characteristics and function study of TNF receptor-associated factor 5 from grouper (Epinephelus coioides). Fish Shellfish Immunol. 2019, 87, 730–736. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.T.; Sun, M.S.; Zhang, X.; Liao, J.M.; Liu, M.K.; Qin, Q.W.; Wei, J.G. Grouper TRAF4, a novel, CP-interacting protein that promotes red-spotted grouper nervous necrosis virus replication. Int. J. Mol. Sci. 2021, 22, 6136. [Google Scholar] [CrossRef]
- Zou, P.F.; Shen, J.J.; Li, Y.; Zhang, Z.P.; Wang, Y.L. TRAF3 enhances TRIF-mediated signaling via NF-κB and IRF3 activation in large yellow croaker Larimichthys crocea. Fish Shellfish Immunol. 2020, 97, 114–124. [Google Scholar] [CrossRef]
- Swaidani, S.; Liu, C.N.; Zhao, J.J.; Bulek, K.; Li, X.X. TRAF Regulation of IL-17 Cytokine Signaling. Front. Immunol. 2019, 10, 1293. [Google Scholar] [CrossRef]
- Xia, H.L.; Li, Y.; Wang, Z.W.; Chen, W.J.; Cheng, J.; Yu, D.P.; Lu, Y.S. Expression and functional analysis of tumor necrosis factor receptor (TNFR)-associated factor 5 from Nile tilapia, Oreochromis niloticus. Fish Shellfish Immunol. 2019, 93, 781–788. [Google Scholar] [CrossRef]
- Clawson, G.A.; Smuckler, E.A. A model for nucleocytoplasmic transport of ribonucleoprotein particles. J. Theor. Biol. 1982, 95, 607–613. [Google Scholar] [CrossRef]
- Rusch, S.L.; Kendall, D.A. Protein transport via amino-terminal targeting sequences: Common themes in diverse systems. Mol. Membr. Biol. 1995, 12, 295–307. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; Heijne, G.V. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence, J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Cokol, M.; Nair, R.; Rost, B. Finding nuclear localization signals. EMBO Rep. 2000, 1, 411–415. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.R.; Rotheman, J.E. Biosynthetic transport and sorting by the endoplasmatic reticulum and Golgi. Annu. Rev. Biochem. 1987, 56, 829–852. [Google Scholar] [CrossRef] [PubMed]
- Rost, B.; Nair, J.L.; Wrzeszczynski, K.P.; Ofran, Y. Automatic prediction of protein function, Cell. Mol. Life Sci. 2003, 60, 2637–2650. [Google Scholar]
- Regnier, C.H.; Tomasetto, C.; Moog-Lutz, C.; Chenard, M.P.; Wendling, C.; Basset, P.; Rio, M.C. Presence of a new conserved domain in CART1, a novel member of the tumor necrosis factor receptor-associated protein family, which is expressed in breast carcinoma. J. Biol. Chem. 1995, 270, 25715–25721. [Google Scholar] [CrossRef]
- Yang, M.; Han, R.; Ni, L.Y.; Luo, X.C.; Li, A.X.; Dan, X.M.; Li, Y.W. Molecular characteristics and functional study of tumor necrosis factor receptor-associated factor 2 from the orange-spotted grouper (Epinephelus coioides). Fish Shellfish Immunol. 2019, 84, 726–732. [Google Scholar] [CrossRef]
- Wu, S.T.; Sun, M.S.; Zhang, L.H.; Kang, S.Z.; Liao, J.M.; Zhu, Z.; Chen, H.; Xu, Z.Q.; Xu, L.T.; Zhang, X.; et al. Grouper TRAF3 inhibits nodavirus infection by regulating the STING-mediated antiviral signaling pathway. Fish Shellfish Immunol. 2022, 123, 172–181. [Google Scholar] [CrossRef]
- Sun, M.S.; Wu, S.T.; Zhang, X.; Zhang, L.H.; Kang, S.Z.; Qin, Q.W.; Wei, J.G. Grouper TRAF5 exerts negative regulation on antiviral immune response against iridovirus. Fish Shellfish Immunol. 2021, 115, 7–13. [Google Scholar] [CrossRef]
- He, J.Q.; Zarnegar, B.; Oganesyan, G.; Saha, S.K.; Yamazaki, S.; Doyle, S.E.; Dempsey, P.W.; Cheng, G.H. Rescue of TRAF3-null mice by p100 NF-kappa B deficiency. J. Exp. Med. 2006, 203, 2413–2418. [Google Scholar] [CrossRef]
- Cabal-Hierro, L.; Rodríguez, M.; Artime, N.; Iglesias, J.; Ugarte, L.; Prado, M.A.; Lazo, P.S. TRAF-mediated modulation of NF-kB and jnk activation by TNFR2. Cell. Signal. 2014, 26, 2658–2666. [Google Scholar] [CrossRef]
- Chen, H.; Xiao, J.; Li, J.; Liu, J.; Wang, C.Y.; Feng, C.L.; Feng, H. TRAF2 of black carp upregulates MAVS-mediated antiviral signaling during innate immune response. Fish Shellfish Immunol. 2017, 71, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Xia, H.L.; Huang, Y.C.; Tang, J.F.; Jian, J.C.; Wu, Z.H.; Lu, Y.S. Identification and characterization of tumor necrosis factor receptor (TNFR)-associated factor 3 from humphead snapper, Lutjanus sanguineus. Fish Shellfish Immunol. 2015, 46, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Kedinger, V.; Rio, M.C. TRAF4, the unique family member. Adv. Exp. Med. Biol. 2007, 597, 60–71. [Google Scholar]
- Wang, X.; Song, X.J.; Xie, X.C.; Li, W.Z.; Lu, L.; Chen, S.; Wu, H.; Feng, H. TRAF3 enhances STING mediated antiviral signaling during the innate immune activation of black carp. Dev. Comp. Immunol. 2018, 88, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.L.; Wang, Z.W.; Yu, D.P.; Xia, L.Q.; Chen, W.J.; Long, M.; Fan, H.M.; Xia, H.L.; Lu, Y.S. Characterization of TRAF2 in Nile tilapia: Expression profiles and the role in decreasing NF-κB pathway. Fish Shellfish Immunol. 2022, 122, 13–20. [Google Scholar] [CrossRef]
- Min, W.; Bradley, J.R.; Galbraith, J.J.; Jones, S.J.; Ledgerwood, E.C.; Pober, J.S. The N-terminal domains target TNF receptor-associated factor-2 to the nucleus and display transcriptional regulatory activity. J. Immunol. 1998, 161, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Zhang, G.R.; Zhu, D.M.; Shi, Z.C.; Liao, C.L.; Fan, Q.X.; Wei, K.J.; Ji, W. Molecular characterization and expression analysis of IL-22 and its two receptors genes in yellow catfish (Pelteobagrus filvidraco) in response to Edwardsiella ictaluri challenge. Fish Shellfish Immunol. 2017, 66, 466–479. [Google Scholar] [CrossRef]
- Söding, J.; Biegert, A.; Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005, 33, W244–W248. [Google Scholar] [CrossRef]
- Hall, B.G. Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhang, G.R.; Ji, W.; Shi, Z.C.; Ma, X.F.; Luo, Z.L.; Wei, K.J. Expression and Function Analysis of interleukin-17A/F1, 2 and 3 Genes in yellow catfish (Pelteobagrus fulvidraco): Distinct bioactivity of recombinant IL-17A/F1, 2, and 3. Front. Immunol. 2021, 12, 626895. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]








| Gene | ORF (bp) | AA | MW (kDa) | pI |
|---|---|---|---|---|
| Pf_TRAF1 | 1629 | 542 | 60.55 | 6.95 |
| Pf_TRAF2a | 1839 | 612 | 69.09 | 7.96 |
| Pf_TRAF2b | 1554 | 517 | 58.76 | 7.90 |
| Pf_TRAF3 | 1671 | 556 | 63.90 | 7.55 |
| Pf_TRAF4a | 1413 | 470 | 54.16 | 7.91 |
| Pf_TRAF4b | 1437 | 478 | 54.97 | 8.26 |
| Pf_TRAF5 | 1653 | 550 | 62.77 | 6.47 |
| Pf_TRAF6 | 1617 | 538 | 61.48 | 6.45 |
| Pf_TRAF7 | 2025 | 674 | 74.86 | 6.64 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
You, S.-L.; Jiang, X.-X.; Zhang, G.-R.; Ji, W.; Ma, X.-F.; Zhou, X.; Wei, K.-J. Molecular Characterization of Nine TRAF Genes in Yellow Catfish (Pelteobagrus fulvidraco) and Their Expression Profiling in Response to Edwardsiella ictaluri Infection. Int. J. Mol. Sci. 2023, 24, 8363. https://doi.org/10.3390/ijms24098363
You S-L, Jiang X-X, Zhang G-R, Ji W, Ma X-F, Zhou X, Wei K-J. Molecular Characterization of Nine TRAF Genes in Yellow Catfish (Pelteobagrus fulvidraco) and Their Expression Profiling in Response to Edwardsiella ictaluri Infection. International Journal of Molecular Sciences. 2023; 24(9):8363. https://doi.org/10.3390/ijms24098363
Chicago/Turabian StyleYou, Shen-Li, Xin-Xin Jiang, Gui-Rong Zhang, Wei Ji, Xu-Fa Ma, Xu Zhou, and Kai-Jian Wei. 2023. "Molecular Characterization of Nine TRAF Genes in Yellow Catfish (Pelteobagrus fulvidraco) and Their Expression Profiling in Response to Edwardsiella ictaluri Infection" International Journal of Molecular Sciences 24, no. 9: 8363. https://doi.org/10.3390/ijms24098363
APA StyleYou, S. -L., Jiang, X. -X., Zhang, G. -R., Ji, W., Ma, X. -F., Zhou, X., & Wei, K. -J. (2023). Molecular Characterization of Nine TRAF Genes in Yellow Catfish (Pelteobagrus fulvidraco) and Their Expression Profiling in Response to Edwardsiella ictaluri Infection. International Journal of Molecular Sciences, 24(9), 8363. https://doi.org/10.3390/ijms24098363







