Molecular Functions and Physiological Roles of Gustatory Receptors of the Silkworm Bombyx mori
Abstract
:1. Introduction
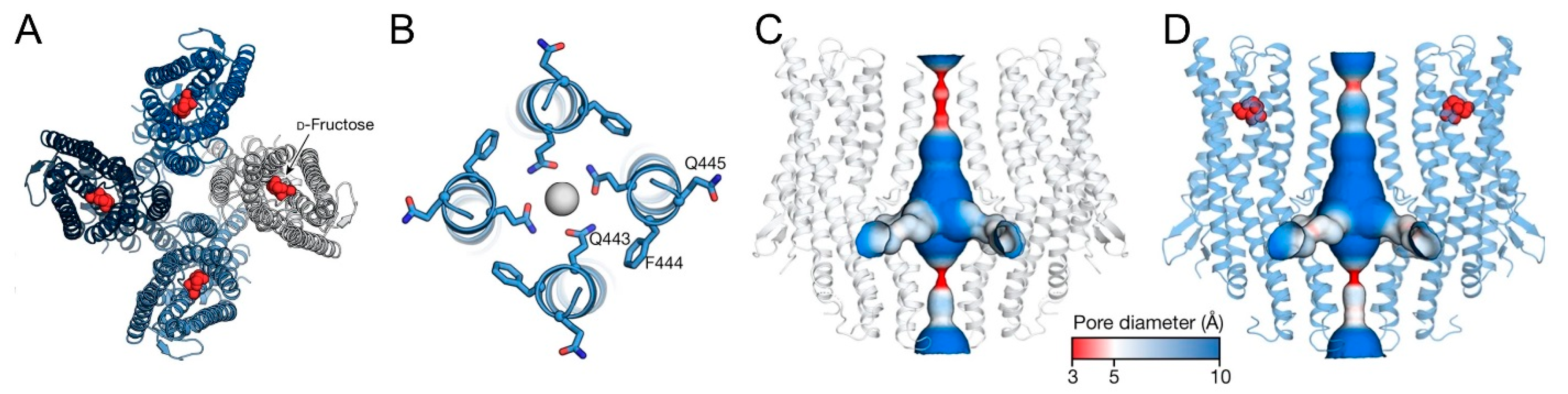
2. Structure- and Ligand-Specific Ion Channel Opening Function of BmGrs
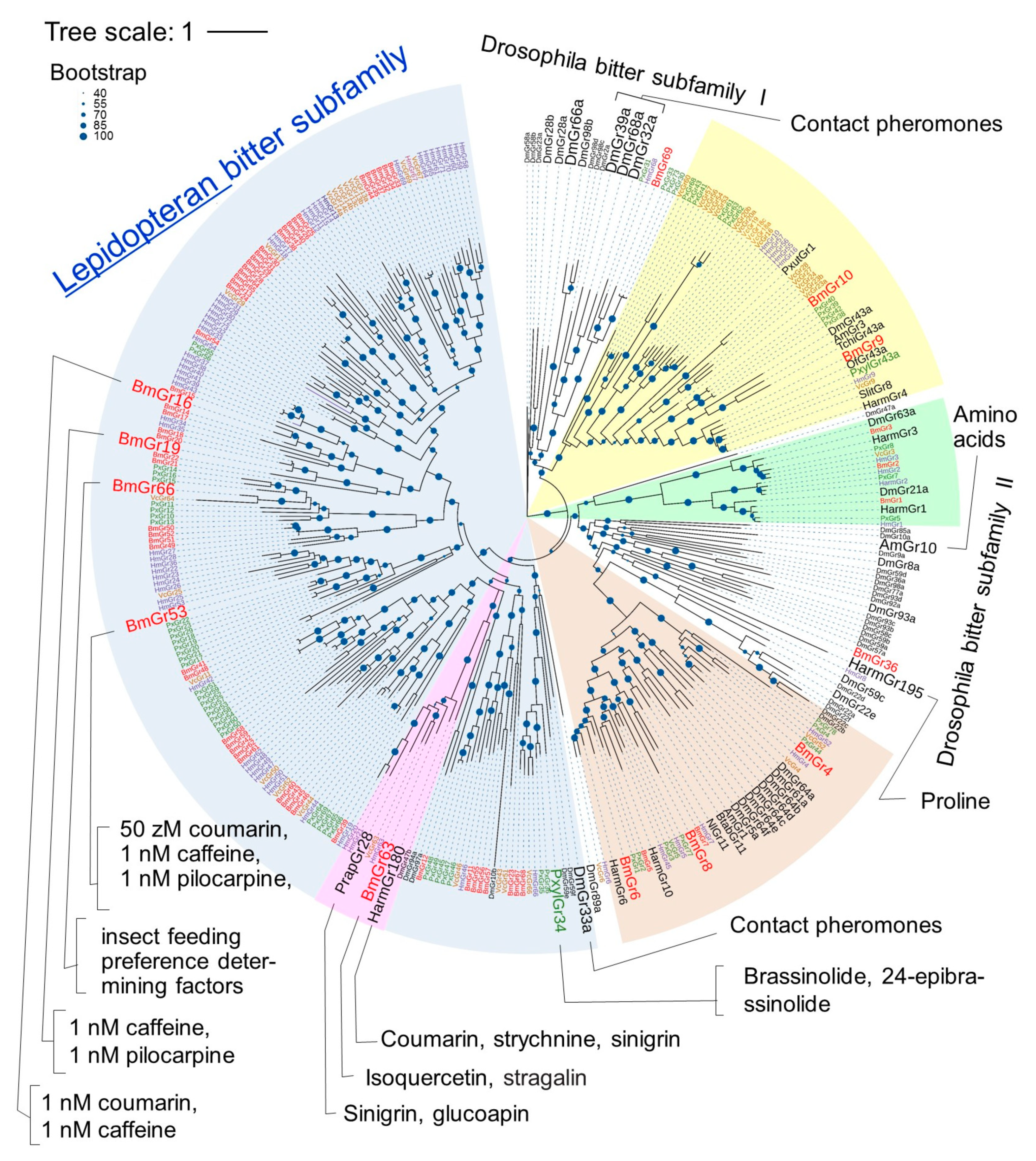
3. Subfamilies Comprising the BmGr Phylogenetic Tree and Ligands of BmGrs
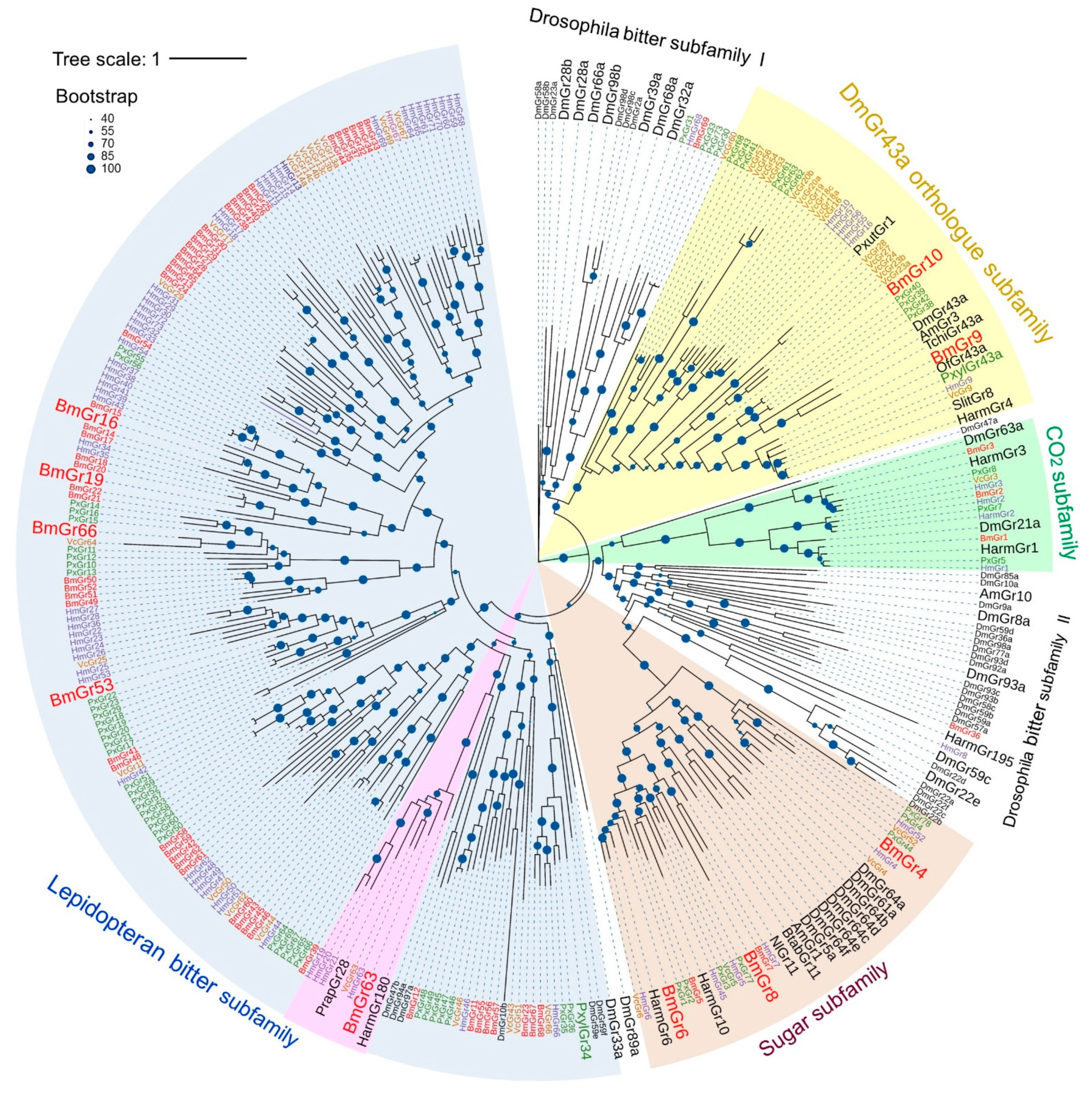
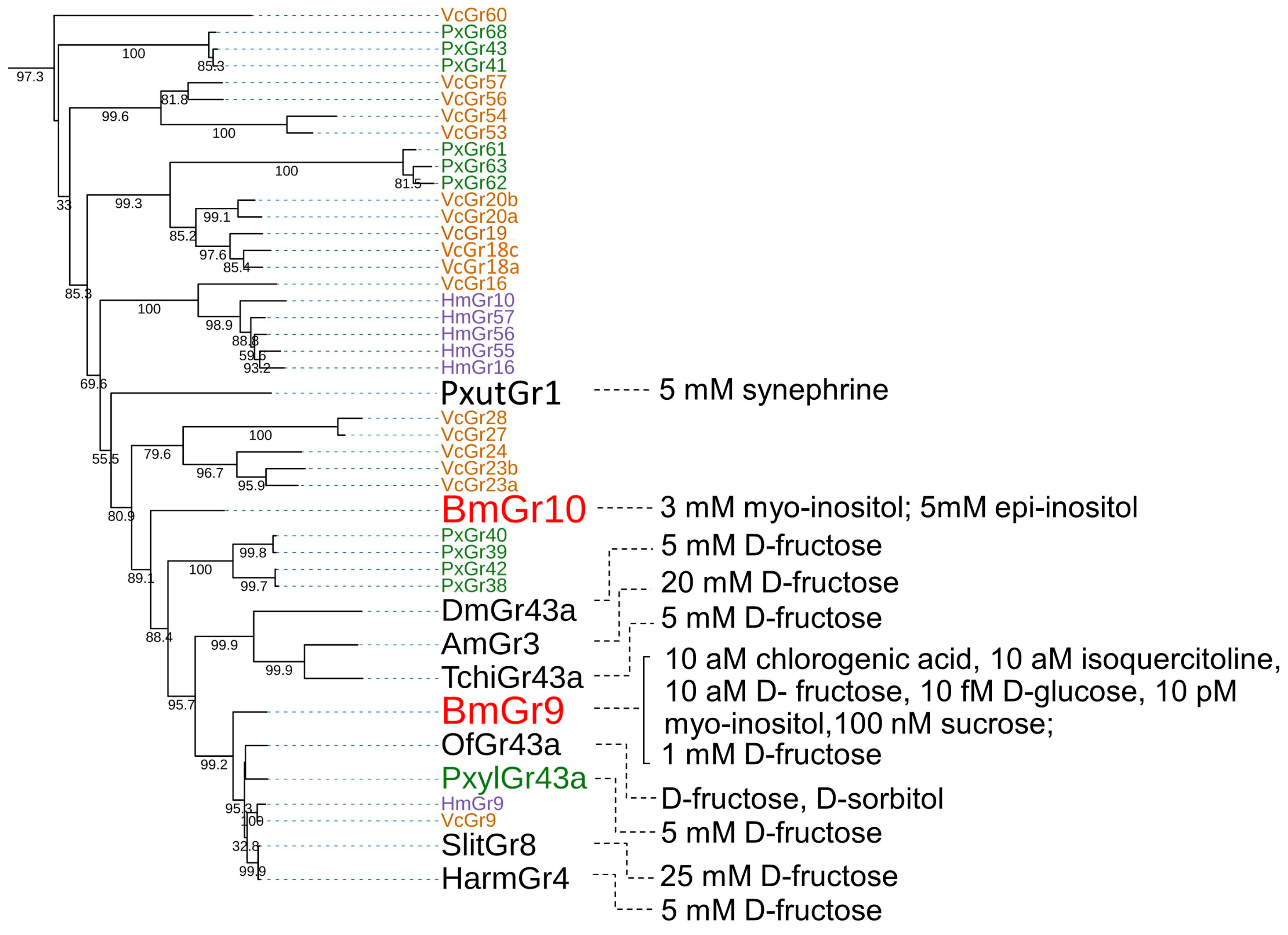
3.1. DmGr43a Orthologue Subfamily
3.1.1. Ligands of BmGr9
3.1.2. Ligands of BmGr10
3.1.3. DmGr43a Orthologue Subfamily Members of the Lepidopteran Insects
3.2. Sugar Subfamily
3.2.1. Ligands of BmGr4
3.2.2. Ligands of BmGr6
3.2.3. Ligands of BmGr8
3.2.4. Sugar Subfamily Members of the Lepidopteran Insects
3.3. Bitter Subfamily
3.3.1. Ligands of BmGr16, BmGr19, and BmGr53
3.3.2. Ligands of Bmgr63
3.3.3. Ligands of BmGr66
3.3.4. Bitter Subfamily Members of Lepidopteran Insects
4. Gr-Expressing Organs and the Roles of Grs
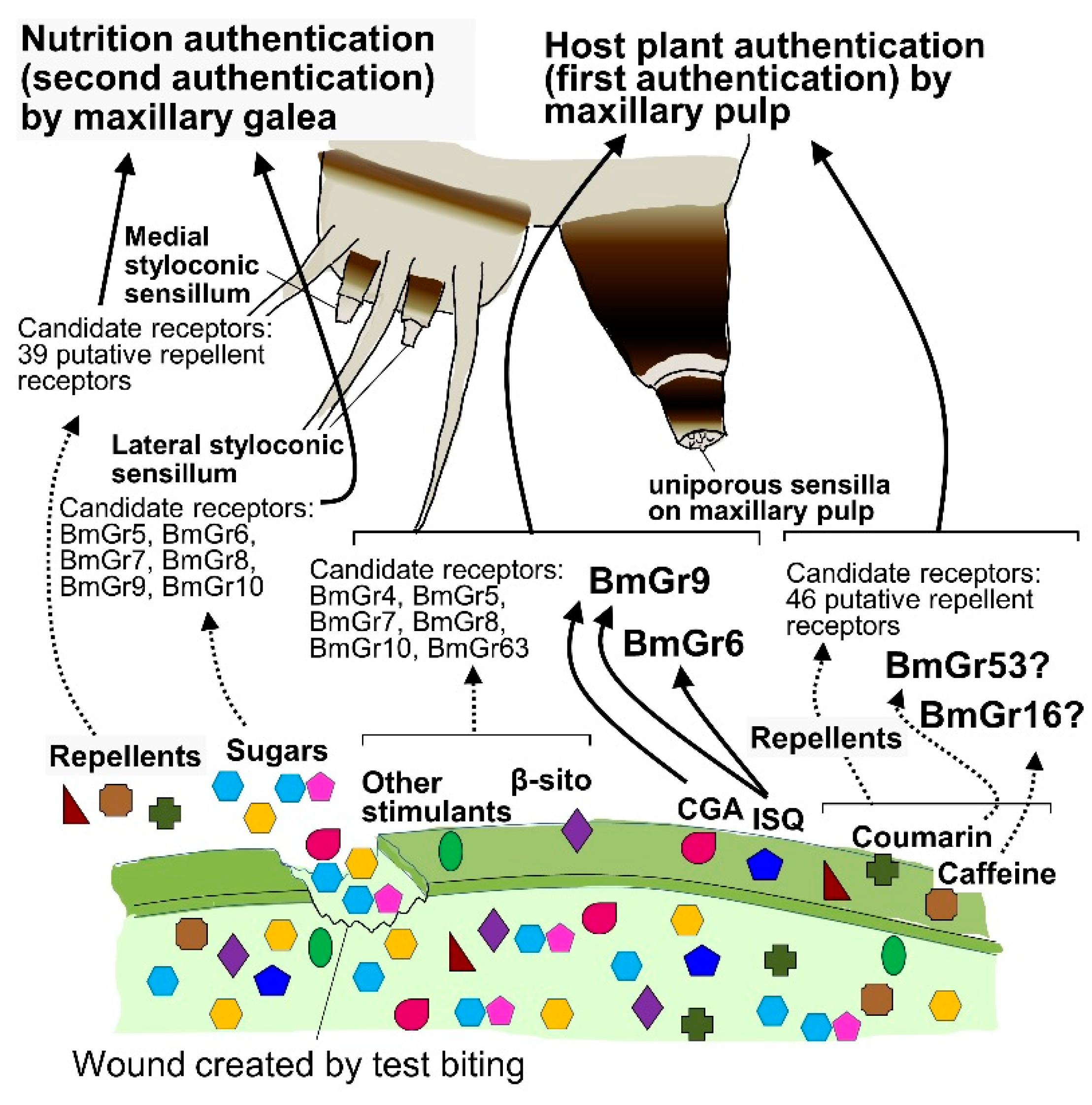
4.1. Spatial Expression of BmGrs
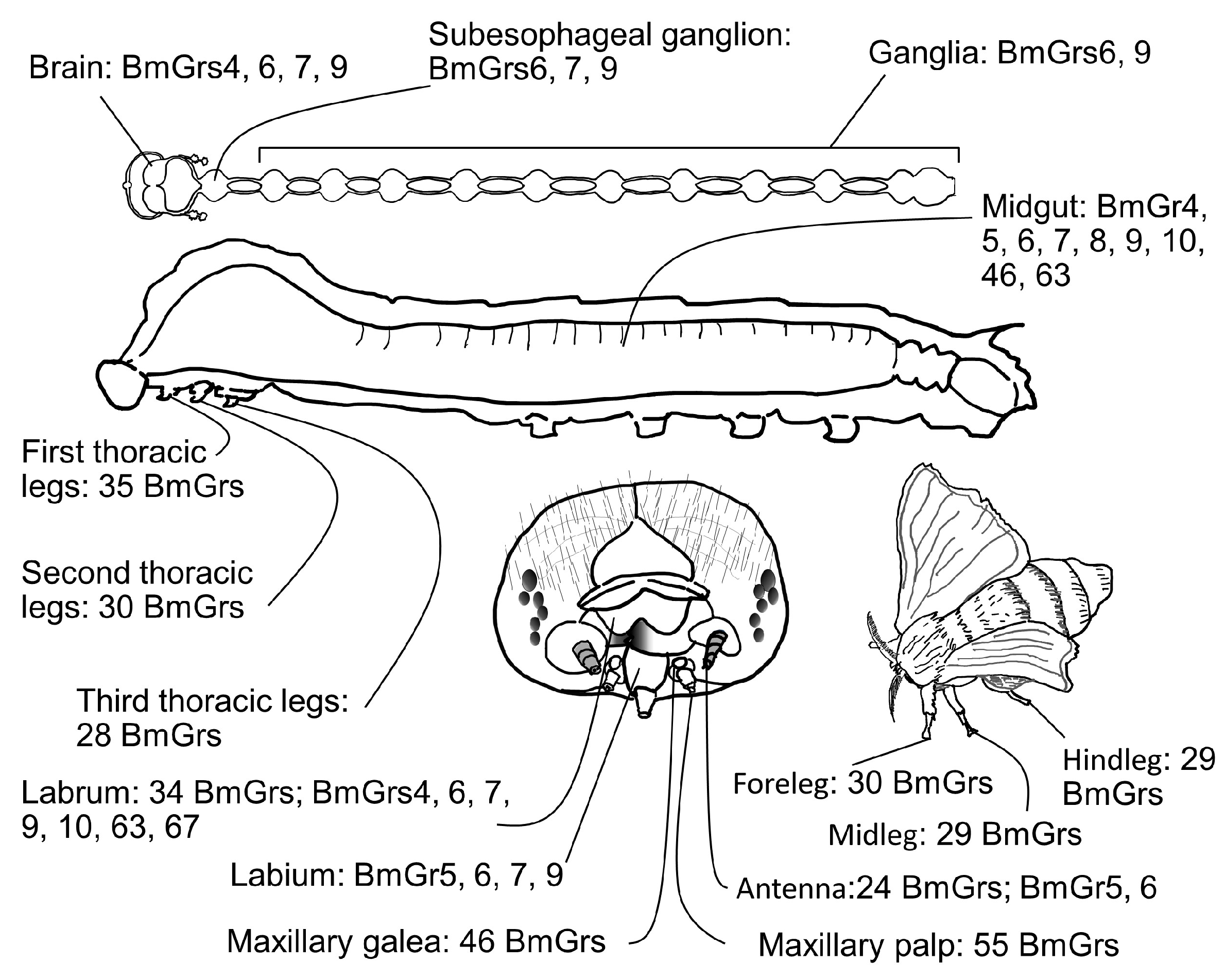
4.2. Roles of BmGr6 and BmGr9 in First Authentication of Food by the Maxillary Palps
4.3. Role of Bitter Receptors in the First Authentication of Food by the Maxillary Palps
4.4. Food Recognition through Midgut Expressed BmGrs
4.4.1. Expression and Role of BmGr4 in Midgut Enteroendocrine Cells
4.4.2. Expression and Role of BmGr6 in Midgut Enteroendocrine Cells
4.4.3. Expression of Other BmGrs in Midgut Enteroendocrine Cells
4.5. Roles of BmGrs beyond Taste Recognition
Funding
Conflicts of Interest
References
- Schoonhoven, L.M.; van Loon, J.J.A.; Dicke, M. Insect-Plant Biology, 2nd ed.; Oxford University Press: Oxford, UK, 2005; 421p. [Google Scholar]
- Nishida, R. Chemosensory basis of host recognition in butterflies—Multi-component system of oviposition stimulants and deterrents. Chem. Senses 2005, 30, i293–i294. [Google Scholar] [CrossRef]
- Nishida, R. Chemical ecology of insect-plant interactions: Ecological significance of plant secondary metabolites. Biosci. Biotechnol. Biochem. 2014, 78, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Anton, S.; Cortesero, A.M. Plasticity in chemical host plant recognition in herbivorous insects and its implication for pest control. Biology 2022, 11, 1842. [Google Scholar] [CrossRef] [PubMed]
- Schoonhoven, L.M.; van Loon, J.J.A. An inventory of taste in caterpillars: Each species its own key. Acta Zool. Acad. Sci. H 2002, 48, 215–263. [Google Scholar]
- Hamamura, Y.; Hayashiya, K.; Naito, K. Food selection by silkworm larvae, Bombyx mori: β-sitosterol as one of the biting factors. Nature 1961, 190, 880–881. [Google Scholar] [CrossRef]
- Hamamura, Y.; Hayashiya, K.; Naito, K.; Matsuura, K.; Nishida, J. Food selection by silkworm larvae. Nature 1962, 194, 754–755. [Google Scholar] [CrossRef]
- Tsuneto, K.; Endo, H.; Shii, F.; Sasaki, K.; Nagata, S.; Sato, R. Diet choice: The two-factor host acceptance system of silkworm larvae. PLoS Biol. 2020, 18, e3000828. [Google Scholar] [CrossRef]
- Ishikawa, S. Responses of maxillary chemoreceptors in the larva of the silkworm, Bombyx mori, to stimulation by carbohydrates. J. Cell. Comp. Physiol. 1963, 61, 99–107. [Google Scholar] [CrossRef]
- Ishikawa, S. Electrical response and function of a bitter substance receptor associated with the maxillary sensilla of the larva of the silkworm, Bombyx mori L. J. Cell. Physiol. 1966, 67, 1–11. [Google Scholar] [CrossRef]
- Asaoka, K.; Shibuya, T. Morphological and electrophysiological characteristics of the epipharyngeal sensilla of the silkworm, Bombyx mori. Entomol. Exp. Appl. 1995, 77, 167–176. [Google Scholar] [CrossRef]
- Yang, J.; Guo, H.; Jiang, N.-J.; Tang, R.; Li, G.-C.; Huang, L.-Q.; van Loon, J.J.A.; Wang, C.-Z. Identification of a gustatory receptor tuned to sinigrin in the cabbage butterfly Pieris rapae. PLoS Genet. 2021, 17, e1009527. [Google Scholar] [CrossRef] [PubMed]
- Chyb, S.; Dahanukar, A.; Wickens, A.; Carlson, J.R. Drosophila Gr5a encodes a taste receptor tuned to trehalose. Proc. Natl. Acad. Sci. USA 2003, 100, 14526–14530. [Google Scholar] [CrossRef] [PubMed]
- Robertson, H.M.; Warr, C.G.; Carlson, J.R. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2003, 100, 14537–14542. [Google Scholar] [CrossRef] [PubMed]
- Wanner, K.W.; Robertson, H.M. The gustatory receptor family in the silkworm moth Bombyx mori is characterized by a large expansion of a single lineage of putative bitter receptors. Insect Mol. Biol. 2008, 17, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-J.; Anderson, A.R.; Trowell, S.C.; Luo, A.-R.; Xiang, Z.-H.; Xia, Q.-Y. Topological and functional characterization of an insect gustatory receptor. PLoS ONE 2011, 6, e24111. [Google Scholar]
- Guo, H.; Cheng, T.; Chen, Z.; Jiang, L.; Guo, Y.; Liu, J.; Li, S.; Taniai, K.; Asaoka, K.; Kadono-Okuda, K.; et al. Expression map of a complete set of gustatory receptor genes in chemosensory organs of Bombyx mori. Insect Biochem. Mol. Biol. 2017, 82, 74–82. [Google Scholar]
- Zhan, S.; Merlin, C.; Boore, J.L.; Reppert, S.M. The monarch butterfly genome yields insights into long-distance migration. Cell 2011, 147, 1171–1185. [Google Scholar] [CrossRef]
- Briscoe, A.D.; Macias-Muñoz, A.; Kozak, K.M.; Walters, J.R.; Yuan, F.; Jamie, G.A.; Martin, S.H.; Dasmahapatra, K.K.; Ferguson, L.C.; Mallet, J.; et al. Female behaviour drives expression and evolution of gustatory receptors in butterflies. PLoS Genet. 2013, 9, e1003620. [Google Scholar] [CrossRef]
- Xu, W.; Papanicolaou, A.; Zhang, H.-J.; Anderson, A. Expansion of a bitter taste receptor family in a polyphagous insect herbivore. Sci. Rep. 2016, 6, 23666. [Google Scholar]
- Suzuki, H.C.; Ozaki, K.; Makino, T.; Uchiyama, H.; Yajima, S.; Kawata, M. Evolution of gustatory receptor gene family provides insights into adaptation to diverse host plants in nymphalid butterflies. Genome Biol. Evol. 2018, 10, 1351–1362. [Google Scholar] [CrossRef]
- Yang, K.; Gong, X.-L.; Li, G.-C.; Huang, L.-Q.; Ning, C.; Wang, C.-Z. A gustatory receptor tuned to the steroid plant hormone brassinolide in Plutella xylostella (Lepidoptera: Plutellidae). eLife 2020, 9, e64114. [Google Scholar] [CrossRef] [PubMed]
- Xu, W. How do moth and butterfly taste?—Molecular basis of gustatory receptors in Lepidoptera insect. Science 2020, 27, 1148–1157. [Google Scholar]
- Aryal, B.; Dhakal, S.; Shrestha, B.; Lee, Y. Molecular and neuronal mechanisms for amino acid taste perception in the Drosophila labellum. Curr. Biol. 2022, 32, 1376–1386.e4. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Tang, J.; Li, Y.; Li, D.; Chen, G.; Chen, L.; Yang, Z.; He, N. The silkworm gustatory receptor BmGr63 is dedicated to the detection of isoquercetin in mulberry. Proc. R. Soc. B 2022, 289, 20221427. [Google Scholar] [CrossRef] [PubMed]
- Endo, H.; Tsuneto, K.; Mang, D.; Zhang, W.; Yamagishi, T.; Ito, K.; Nagata, S.; Sato, R. Molecular basis of host plant recognition by silkworm larvae. J. Insect Physiol. 2024, 154, 104628. [Google Scholar] [CrossRef]
- Shrestha, B.; Aryal, B.; Lee, Y. The taste of vitamin C in Drosophila. EMBO Rep. 2023, 24, e56319. [Google Scholar] [CrossRef]
- Mang, D.; Shu, M.; Endo, H.; Yoshizawa, Y.; Nagata, S.; Kikuta, S.; Sato, R. Expression of a sugar clade gustatory receptor, BmGr6, in the oral sensory organs, midgut, and central nervous system of larvae of the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 2016, 70, 85–98. [Google Scholar] [CrossRef]
- Mang, D.; Shu, M.; Tanaka, S.; Nagata, S.; Takada, T.; Endo, H.; Kikuta, S.; Tabunoki, H.; Iwabuchi, K.; Sato, R. Expression of the fructose receptor BmGr9 and its involvement in the promotion of feeding, suggested by its co-expression with neuropeptide F1 in Bombyx mori. Insect Biochem. Mol. Biol. 2016, 75, 58–69. [Google Scholar] [CrossRef]
- Mang, D.; Mayu, K.; Toyama, T.; Yamagishi, T.; Sato, R. BmGr4 responds to sucrose and glucose and expresses in tachykinin-related peptid-secreting enteroendocrine cells. Insect Biochem. Mol. Biol. 2022, 150, 103858. [Google Scholar] [CrossRef]
- Toyam, T.; Yamagishi, T.; Sato, R. The roles of enteroendocrine cell distribution and gustatory receptor expression in regulating peptide hormone secretion in the midgut of Bombyx mori larvae. Arch. Insect Biochem. Physiol. 2023, 114, e22032. [Google Scholar] [CrossRef]
- Mang, D.; Toyama, T.; Yamagishi, T.; Sun, J.; Purba, E.R.; Endo, H.; Matthews, M.M.; Ito, K.; Nagata, S.; Sato, R. Dietary compounds activate an insect gustatory receptor on enteroendocrine cells to elicit myosuppressin secretion. Insect Biochem. Mol. Biol. 2023, 155, 103927. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Tanaka, K.; Touhara, K. Sugar-regulated cation channel formed by an insect gustatory receptor. Proc. Natl. Acad. Sci. USA 2011, 108, 11680–11685. [Google Scholar] [CrossRef] [PubMed]
- Kikuta, S.; Endo, H.; Tomita, N.; Takada, T.; Morita, C.; Asaoka, K.; Sato, R. Characterization of a ligand-gated cation channel based on an inositol receptor in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2016, 74, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Frank, H.M.; Walujkar, S.; Walsh, R.M., Jr.; Laursen, W.J.; Theobald, D.L.; Garrity, P.A.; Gaudet, R. Structural basis of ligand specificity and channel activation in an insect gustatory receptor. Cell Rep. 2024, 43, 114035. [Google Scholar] [CrossRef]
- Gomes, J.V.; Singh-Bhagania, S.; Cenci, M.; Cordon, C.C.; Singh, M.; Butterwick, J.A. Structural basis of ligand specificity and channel activation in an insect gustatory receptor. Nature 2024, 629, 228–234. [Google Scholar] [CrossRef]
- Jones, W.D.; Cayirlioglu, P.; Kadow, I.G.; Vosshall, L.B. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature 2007, 445, 86–90. [Google Scholar] [CrossRef]
- Jiao, Y.; Moon, S.J.; Wang, X.; Ren, Q.; Montell, C. Gr64f is required in combination with other gustatory receptors for sugar detection in Drosophila. Curr. Biol. 2008, 18, 1797–1801. [Google Scholar] [CrossRef]
- Lee, Y.; Moon, S.J.; Montell, C. Multiple gustatory receptors required for the caffeine response in Drosophila. Proc. Natl. Acad. Sci. USA 2009, 106, 4495–4500. [Google Scholar] [CrossRef]
- Erdelyan, C.N.G.; Mahood, T.H.; Bader, T.S.Y.; Whyard, S. Functional validation of the carbon dioxide receptor genes in Aedes aegypti mosquitoes using RNA interference. Insect Mol. Biol. 2012, 21, 119–127. [Google Scholar] [CrossRef]
- Ning, C.; Yang, K.; Xu, M.; Huang, L.-Q.; Wang, C.-Z. Functional validation of the carbon dioxide receptor in labial palps of Helicoverpa armigera moths. Insect Biochem. Mol. Biol. 2016, 73, 12–19. [Google Scholar] [CrossRef]
- Elmore, T.; Ignell, R.; Carlson, J.R.; Smith, D.P. Targeted mutation of a Drosophila odor receptor defines receptor requirement in a novel class of sensillum. J. Neurosci. 2003, 23, 9906–9912. [Google Scholar] [CrossRef] [PubMed]
- Larsson, M.C.; Domingos, A.I.; Jones, W.D.; Chiappe, M.E.; Amrein, H.; Vosshall, L.B. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 2004, 43, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; He, L.; Du, J.; Wang, C.-Z.; Zhao, Z. Mechanism of foraging selections regulated by gustatory receptor 43a in Ostrinia furnacalis larvae. Pest Manag. Sci. 2024, 80, 978–987. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-L.; Yan, Q.; Yang, Y.-L.; Hou, W.; Miao, C.-L.; Peng, Y.-C.; Dong, S.-L. A gustatory receptor GR8 tunes specifically to D-fructose in the common cutworm Spodoptera litura. Insects 2019, 10, 272. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wu, H.; Yi, J.; Jiang, D.; Zhang, G. Identification and functional characterization of D-fructose receptor in an egg parasitoid, Trichogramma chilonis. PLoS ONE 2019, 14, e0217493. [Google Scholar] [CrossRef]
- Chen, W.-W.; Kang, K.; Yang, P.; Zhang, W.-Q. Identification of a sugar gustatory receptor and its effect on fecundity of the brown planthopper Nilaparvata lugens. Insect Sci. 2019, 26, 441–452. [Google Scholar] [CrossRef]
- Ozaki, K.; Ryuda, M.; Yamada, A.; Utoguchi, A.; Ishimoto, H.; Calas, D.; Marion-Poll, F.; Tanimura, T.; Yoshikawa, H. A gustatory receptor involved in host plant recognition for oviposition of a swallowtail butterfly. Nat. Commun. 2011, 2, 542. [Google Scholar] [CrossRef]
- Robertson, H.M.; Wanner, K.W. The chemoreceptor superfamily in the honey bee, Apis mellifera: Expansion of the odorant, but not gustatory, receptor family. Genome Res. 2006, 16, 1395–1403. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-tree: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Miyamoto, T.; Slone, J.; Song, X.; Amrein, H. A fructose receptor functions as a nutrient sensor in the Drosophila brain. Cell 2012, 151, 1113–1125. [Google Scholar] [CrossRef] [PubMed]
- Takada, T.; Sasaki, T.; Sato, R.; Kikuta, S.; Inoue, M.N. Differential expression of a fructose receptor gene in honey bee workers according to age and behavioral role. Arch. Insect Biochem. Physiol. 2018, 97, e21437. [Google Scholar] [CrossRef] [PubMed]
- Değirmenci, L.; Geiger, D.; Rogé, F.F.L.; Keller, A.; Krischke, B.; Beye, M.; Steffan-Dewenter, I.; Scheiner, R. CRISPR/Cas 9-mediated mutations as a new tool for studying taste in honeybees. Chem. Senses 2020, 45, 655–666. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-L.; Sun, S.-J.; Hou, W.; Zhang, J.; Yan, Q.; Dong, S.-L. Functional characterization of two spliced variants of fructose gustatory receptor in the diamondback moth, Plutella xylostella. Pestic. Biochem. Physiol. 2020, 164, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.-J.; Ning, C.; Guo, H.; Jia, Y.-Y.; Huang, L.-Q.; Qu, M.-J.; Wang, C.-Z. A gustatory receptor tuned to D-fructose in antennal Sensilla chaetica of Helicoverpa armigera. Insect Biochem. Mol. Biol. 2015, 60, 39–46. [Google Scholar] [CrossRef]
- Xu, W.; Zhang, H.J.; Anderson, A. A sugar gustatory receptor identified from the foregut of cotton bollworm Helicoverpa armigera. J. Chem. Ecol. 2012, 38, 1513–1520. [Google Scholar] [CrossRef]
- Shii, F.; Mang, D.; Kasubuchi, M.; Tsuneto, K.; Toyama, T.; Endo, H.; Sasaki, K.; Sato, R. Ultrasensitive detection by maxillary palp neurons allows non-host recognition without consumption of harmful allelochemicals. J. Insect Physiol. 2021, 132, 104263. [Google Scholar] [CrossRef]
- Kasubuchi, M.; Shii, F.; Tsuneto, K.; Yamagishi, T.; Adegawa, S.; Endo, H.; Sato, R. Insect taste receptors relevant to host identification by recognition of secondary metabolite patterns of non-host plants. Biochem. Biophys. Res. Commun. 2018, 499, 901–906. [Google Scholar] [CrossRef]
- Ohsugi, T.; Nishida, R.; Fukami, H. Multi-conmponent system of oviposition stimulants for a Rutaceae-feeding swallowtail butterfly, Papilio xuthus (Lepidoptera: Papilionidae). Appl. Entomol. Zool. 1991, 26, 29–40. [Google Scholar] [CrossRef]
- Nishida, R.; Fukami, H. Oviposition stimulants of an Aristolochiaceae-feeding swallowtail butterfly, Atrophaneura alcinous. J. Chem. Ecol. 1989, 15, 2565–2575. [Google Scholar] [CrossRef]
- Zhang, S.-S.; Wanga, P.-C.; Ninga, C.; Yanga, K.; Lia, G.-C.; Cao, L.-L.; Huanga, L.-Q.; Wanga, C.-Z. The larva and adult of Helicoverpa armigera use differential gustatory receptors to sense sugars. eLife 2024, 12, RP91711. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Moon, S.J.; Montell, C. A Drosophila gustatory receptor required for the responses to sucrose, glucose, and maltose identified by mRNA tagging. Proc. Natl. Acad. Sci. USA 2007, 104, 14110–14115. [Google Scholar] [CrossRef] [PubMed]
- Dahanukar, A.; Lei, Y.-T.; Kwon, J.Y.; Carlson, J.R. Two Gr genes underlie sugar reception in Drosophila neuron. Neuron 2007, 56, 503–516. [Google Scholar] [CrossRef] [PubMed]
- Fujii, S.; Yavuz, A.; Slone, J.; Jagge, C.; Song, X.; Amrein, H. Drosophila sugar receptors in sweet taste perception, olfaction, and internal nutrient sensing. Curr. Biol. 2015, 25, 621–627. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, T.; Chen, Y.; Slone, J.; Amrein, H. Identification of a Drosophila glucose receptor using Ca 2+ imaging of single chemosensory neurons. PLoS ONE 2013, 8, e56304. [Google Scholar] [CrossRef]
- Jung, J.W.; Park, K.W.; Ahn, Y.-J.; Kwon, H.W. Functional characterization of sugar receptors in the western honeybee, Apis mellifera. J. Asia-Pac. Entomol. 2015, 18, 19–26. [Google Scholar] [CrossRef]
- Li, F.; Di, Z.; Tian, J.; Dewer, Y.; Qu, C.; Yang, S.; Luo, C. Silencing the gustatory receptor BtGR11 affects the sensing of sucrose in the whitefly Bemisia tabaci. Front. Bioeng. Biotechnol. 2022, 10, 1054943. [Google Scholar] [CrossRef]
- Hirao, T.; Arai, N. Gustatory and feeding responses to amino acid in the silkworm, Bombyx mori L. J. Appl. Entomol. Zool. 1990, 34, 73–76. [Google Scholar] [CrossRef]
- Zhang, Y.-F.; van Loon, J.J.A.; Wang, C.-Z. Neuron activity and proboscis extension reflex in response to sugars and amino acids in Helicoverpa armigera (Hübner). J. Exp. Biol. 2010, 213, 2889–2895. [Google Scholar] [CrossRef]
- Zhang, Z.-J.; Zhang, S.-S.; Niu, B.-L.; Ji, D.-F.; Liu, X.-J.; Li, M.-W.; Bai, H.; Palli, S.R.; Wang, C.-Z.; Tan, A.-J. A determining factor for insect feeding preference in the silkworm, Bombyx mori. PLoS Biol. 2019, 17, e3000162. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, P.-C.; Zhang, S.-S.; Yang, J.; Li, G.-C.; Huang, L.-Q.; Wang, C.-Z. Functional analysis of a bitter gustatory receptor highly expressed in the larval maxillary galea of Helicoverpa armigera. PLoS Genet. 2022, 18, e1010455. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Jung, J.; Yunusbaev, U.; Ilyasov, R.; Kwon, H.W. Characterization and its implication of a novel taste receptor detecting nutrients in the honey bee, Apis mellifera. Sci. Rep. 2019, 9, 11620. [Google Scholar]
- Giarratani, L.; Vosshall, L.B. Toward a molecular description of pheromone perception. Neuron 2003, 39, 881–883. [Google Scholar] [CrossRef] [PubMed]
- Bray, S.; Amrein, H. A putative Drosophila pheromone receptor expressed in male-specific taste neurons is required for efficient courtship. Neuron 2003, 39, 1019–1029. [Google Scholar] [CrossRef]
- Hu, Y.; Han, Y.; Shao, Y.; Wang, X.; Ma, Y.; Ling, E.; Xue, L. Gr33a modulates Drosophila male courtship preference. Sci. Rep. 2015, 5, 7777. [Google Scholar] [CrossRef]
- Agnihotri, A.R.; Roy, A.A.; Joshi, R.S. Gustatory receptors in Lepidoptera: Chemosensation and beyond. Insect Mol. Biol. 2016, 25, 519–529. [Google Scholar] [CrossRef]
- Shim, J.; Lee, Y.; Jeong, Y.T.; Kim, Y.; Lee, M.G.; Montell, C.; Moonm, S.J. The full repertoire of Drosophila gustatory receptors for detecting an aversive compound. Nat. Commun. 2015, 6, 8867. [Google Scholar] [CrossRef]
- Sung, H.Y.; Jeong, Y.T.; Lim, J.Y.; Kim, H.; Oh, S.M.; Hwang, S.W.; Kwon, J.Y.; Moon, S.J. Heterogeneity in the Drosophila gustatory receptor complexes that detect aversive compounds. Nat. Commun. 2017, 8, 1484. [Google Scholar] [CrossRef]
- Dweck, H.K.M.; Carlson, J.R. Molecular logic and evolution of bitter taste in Drosophila. Curr. Biol. 2020, 30, 17–30.e3. [Google Scholar] [CrossRef]
- Ahn, J.-E.; Amrein, H. Opposing chemosensory functions of closely related gustatory receptors. eLife 2023, 12, RP89795. [Google Scholar] [CrossRef]
- Renwick, J.A.A.; Radke, C.D.; Sachdev-Gupta, K.; Städler, E. Leaf surface chemicals stimulating oviposition by Pieris rapae (Lepidoptera: Pieridae) on cabbage. Chemoecology 1992, 3, 33–38. [Google Scholar] [CrossRef]
- Städler, E.; Renwick, J.A.A.; Radke, C.D.; Sachdev-Gupta, K. Tarsal contact chemoreceptor response to glucosinolates and cardenolides mediating oviposition in Pieris rapae. Physiol. Entomol. 1995, 20, 175–187. [Google Scholar] [CrossRef]
- Renwick, J.A.A.; Lopez, K. Experience-based food consumption by larvae of Pieris rapae: Addiction to glucosinolates? Entomol. Exp. Appl. 1999, 91, 51–58. [Google Scholar] [CrossRef]
- Shields, V.D.C.; Mitchell, B.K. Sinigrin as a feeding deterrent in two crucifer-feeding, polyphagous lepidopterous species and the effects of feeding stimulant mixtures on deterrency. Philos. Trans. R. Soc. B Biol. Sci. 1995, 347, 439–446. [Google Scholar]
- Bernays, E.; Oppenheim, S.; Chapman, R.; Kwon, H.; Gould, F. Taste sensitivity of insect herbivores to deterrents is greater in specialists than in generalists: A behavioral test of the hypothesis with two closely related caterpillars. J. Chem. Ecol. 2000, 26, 547–563. [Google Scholar] [CrossRef]
- Carter, C.; Shafir, S.; Yehonatan, L.; Palmer, R.G.; Thornburg, R. A novel role for proline in plant floral nectars. Naturwissenschaften 2006, 93, 72–79. [Google Scholar] [CrossRef]
- Paerhati, Y.; Ishiguro, S.; Ueda-Matsuo, R.; Yang, P.; Yamashita, T.; Ito, K.; Maekawa, H.; Tani, H.; Suzuki, K. Expression of AmGR10 of the gustatory receptor family in honey bee Is correlated with nursing behavior. PLoS ONE 2015, 10, e0142917. [Google Scholar] [CrossRef]
- Ito, T. Effect of sugars on feedicng of the silkworm, Bombyx mori. J. Insect Physiol. 1960, 5, 95–107. [Google Scholar] [CrossRef]
- Tsuneto, K.; Takada, T.; Kasubuchi, M.; Yamagishi, T.; Adegawa, S.; Sato, R. BmGr10 is a putative functional gustatory receptor in the myo-inositol neuron in the epipharyngeal sensillum. J. Insect Biotechnol. Sericol. 2019, 88, 7–15. [Google Scholar]
- Cheng, D.; Mulder, P.P.J.; van der Meijden, E.; Klinkhamerm, P.G.L.; Vrieling, K. The correlation between leaf-surface and leaf-tissue secondary metabolites: A case study with pyrrolizidine alkaloids in Jacobaea hybrid plants. Metabolomics 2017, 13, 47. [Google Scholar] [CrossRef]
- Tanaka, K.; Uda, Y.; Ono, Y.; Nakagawa, A.; Suwa, M.; Yamaoka, R.; Touhara, K. Highly selective tuning of a silkworm olfactory receptor to a key mulberry leaf volatile. Curr. Biol. 2009, 19, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Glendinning, J.I.; Valcic, S.; Timmermann, B.N. Maxillary palps can mediate taste rejection of plant allelochemicals by caterpillars. J. Comp. Physiol. 1998, 183, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Nagata, S.; Morooka, N.; Matsumoto, S.; Kawai, T.; Nagasawa, H. Effects of neuropeptides on feeding initiation in larvae of the silkworm, Bombyx mori. Gen. Comp. Endocrinol. 2011, 172, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Veenstra, J.A.; Perrimon, N. Control of lipid metabolism by tachykinin in Drosophila. Cell Rep. 2014, 9, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Yamagishia, T.; Endo, H.; Fukumura, K.; Nagata, S.; Hayakawa, T.; Adegawa, S.; Kasubuchi, M.; Sato, R. Glucose, some amino acids and a plant secondary metabolite, chlorogenic acid induce the secretion of a regulatory hormone, tachykinin-related peptide, from the silkworm midgut. Peptides 2018, 106, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Vilaplana, L.; Pascual, N.; Perera, N.; Leira, D.; Bell’es, X. Antifeeding properties of myosuppressin in a generalist phytophagous leafworm, Spodoptera littoralis (Boisduval). Regul. Pept. 2008, 148, 68–75. [Google Scholar] [CrossRef]
- Du, E.J.; Ahn, T.J.; Kwon, I.; Lee, J.H.; Park, J.-H.; Park, S.H.; Kang, T.M.; Cho, H.; Kim, T.J.; Kim, H.-W.; et al. TrpA1 regulates defecation of food-borne pathogens under the control of the Duox pathway. PLoS Genet. 2016, 12, e1005773. [Google Scholar] [CrossRef]
- Roller, L.; Yamanaka, N.; Watanabe, K.; Daubnerová, I.; Zitnan, D.; Kataoka, H.; Tanaka, Y. The unique evolution of neuropeptide genes in the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 2008, 38, 1147–1157. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sato, R. Molecular Functions and Physiological Roles of Gustatory Receptors of the Silkworm Bombyx mori. Int. J. Mol. Sci. 2024, 25, 10157. https://doi.org/10.3390/ijms251810157
Sato R. Molecular Functions and Physiological Roles of Gustatory Receptors of the Silkworm Bombyx mori. International Journal of Molecular Sciences. 2024; 25(18):10157. https://doi.org/10.3390/ijms251810157
Chicago/Turabian StyleSato, Ryoichi. 2024. "Molecular Functions and Physiological Roles of Gustatory Receptors of the Silkworm Bombyx mori" International Journal of Molecular Sciences 25, no. 18: 10157. https://doi.org/10.3390/ijms251810157






