Effective Field Collection of Pezizales Ascospores for Procuring Diverse Fungal Isolates
Abstract
:1. Introduction
2. Materials and Methods
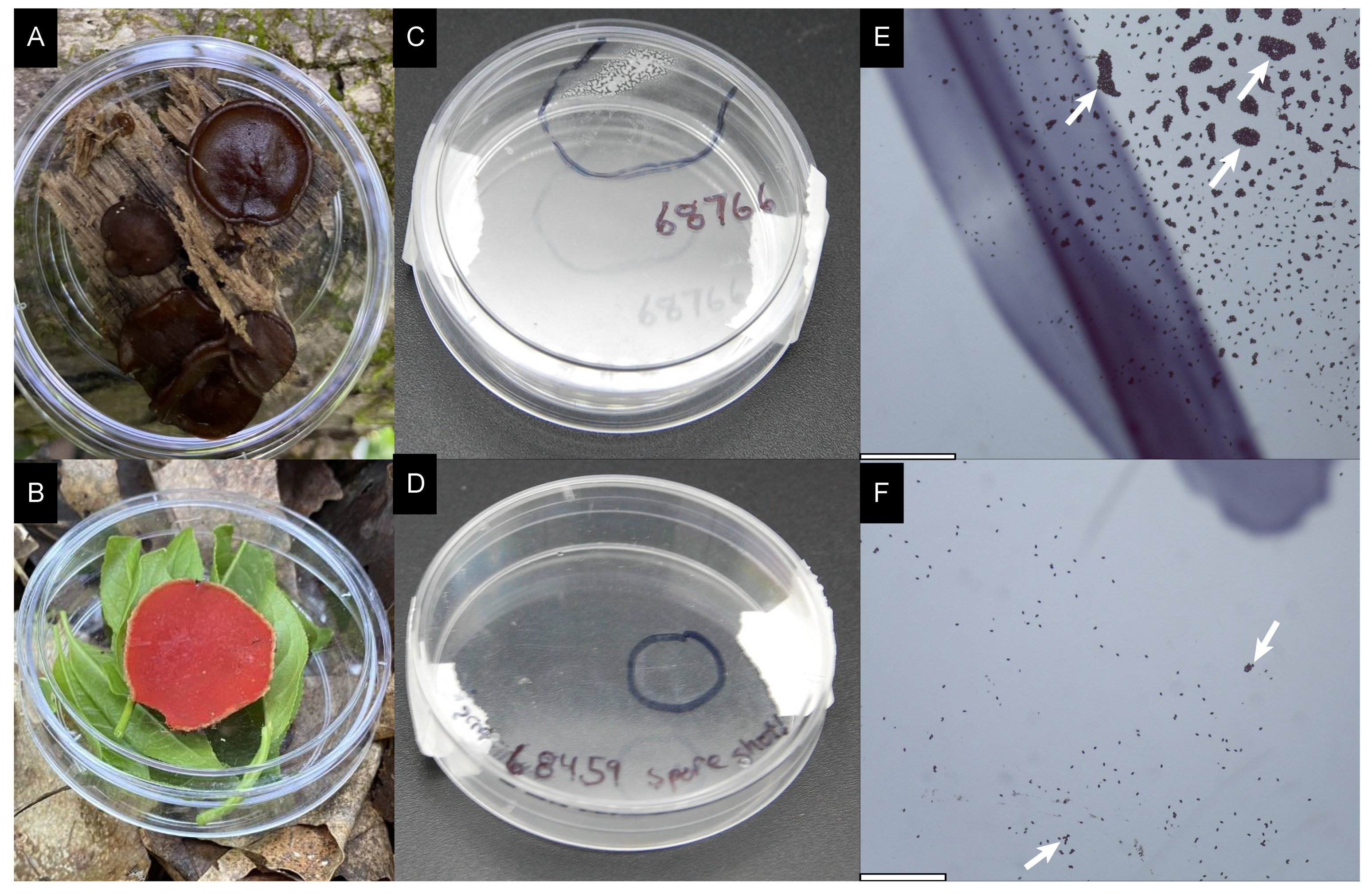
2.1. Collecting Pezizales
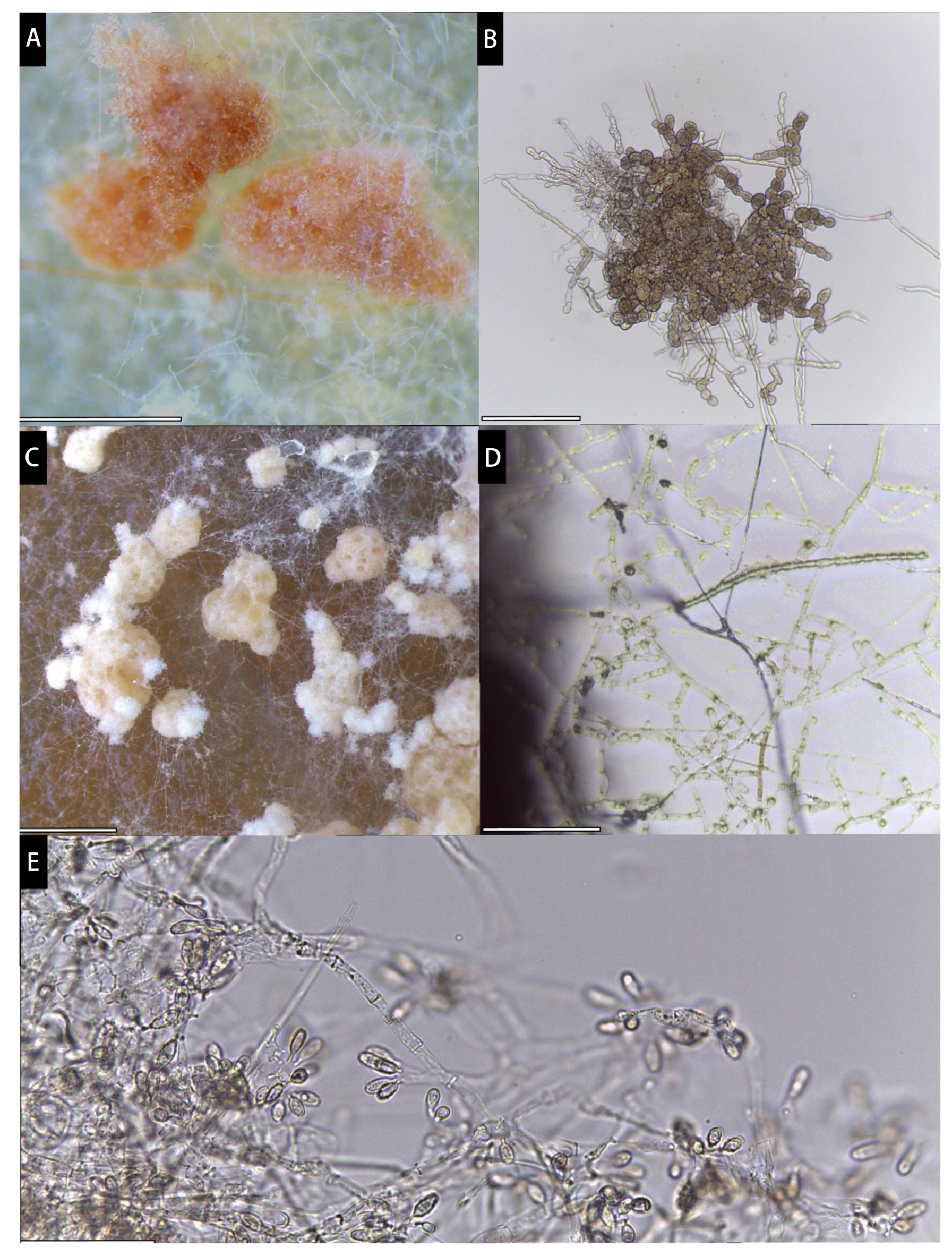
2.2. Germinating Ascospores
2.3. Culture Preservation and Storage
2.4. Visualization and Microscopy
2.5. Molecular Methods
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pfister, D.H.; Healy, R. Pezizomycetes. Encycl. Mycol. 2021, 1, 295–309. [Google Scholar]
- Healy, R.A.; Arnold, A.E.; Bonito, G.; Huang, Y.; Lemmond, B.; Pfister, D.H.; Smith, M.E. Endophytism and endolichenism in Pezizomycetes: The exception or the rule? New Phytol. 2021, 233, 1974–1983. [Google Scholar] [CrossRef]
- Murat, C.; Payen, T.; Noel, B.; Kuo, A.; Morin, E.; Chen, J.; Kohler, A.; Krizsán, K.; Balestrini, R.; Da Silva, C.; et al. Pezizomycetes genomes reveal the molecular basis of ectomycorrhizal truffle lifestyle. Nat. Ecol. Evol. 2018, 2, 1956–1965. [Google Scholar] [CrossRef]
- Pion, M.; Spangenberg, J.E.; Simon, A.; Bindschedler, S.; Flury, C.; Chatelain, A.; Bshary, R.; Job, D.; Junier, P. Bacterial farming by the fungus Morchella crassipes. Proc. Biol. Sci. 2013, 280, 20132242. [Google Scholar]
- Steindorff, A.S.; Seong, K.; Carver, A.; Calhoun, S.; Fischer, M.S.; Stillman, K.; Liu, H.; Drula, E.; Henrissat, B.; Simpson, H.J.; et al. Diversity of genomic adaptations to the post-fire environment in Pezizales fungi points to crosstalk between charcoal tolerance and sexual development. New Phytol. 2022, 236, 1154–1167. [Google Scholar] [CrossRef]
- Miller, A.N.; Bates, S.T. The Mycology Collections Portal (MyCoPortal). IMA Fungus 2017, 8, A65–A66. [Google Scholar] [CrossRef]
- Costantin, J. La culture de la morille et sa forme conidienne. Ann. Sci. Nat. Bot. Series 1936, 18, 111–129. [Google Scholar]
- Paden, J.W. Imperfect states and the taxonomy of the Pezizales. Pers. Mol. Phylogeny Evol. Fungi 1972, 6, 405–414. [Google Scholar]
- Yang, C.S.; Korf, R.P. Ascorhizoctonia gen. nov. and Complexipes emend., two genera for anamorphs of species assigned to Tricharina (Discomycetes). Mycotaxon 1985, 23, 457–481. [Google Scholar]
- Ower, R. Notes on the Development of the Morel Ascocarp: Morchella esculenta. Mycologia 1982, 74, 142–144. [Google Scholar] [CrossRef]
- Kanwal, H.K.; Sudhakara Reddy, M. The effect of carbon and nitrogen sources on the formation of sclerotia in Morchella spp. Ann. Microbiol. 2012, 62, 165–168. [Google Scholar] [CrossRef]
- Liu, W.; Cai, Y.; He, P.; Chen, L.; Bian, Y. Comparative transcriptomics reveals potential genes involved in the vegetative growth of Morchella importuna. 3 Biotech 2019, 9, 81. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yang, J.; Wang, H.; Jin, Y.; Liu, J.; Jia, R.; Wang, Z.; Kang, Z. Analysis of primary metabolites of Morchella fruit bodies and mycelium based on widely targeted metabolomics. Arch. Microbiol. 2021, 204, 98. [Google Scholar] [CrossRef] [PubMed]
- Iotti, M.; Amicucci, A.; Stocchi, V.; Zambonelli, A. Morphological and molecular characterization of mycelia of some Tuber species in pure culture. New Phytol. 2002, 155, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Pfister, D.H. Notes on Caribbean Discomycetes, I.I.I. Ascospore Germination and Growth in Culture of Nanoscypha Tetraspora (Pezizales, Sarcoscyphineae). Mycologia 1973, 65, 952–956. [Google Scholar] [CrossRef]
- Weber, N.S. Western American Pezizales. Selenaspora guernisacii, new to North America. Mycologia 1995, 87, 90–95. [Google Scholar] [CrossRef]
- Volk, T.; Leonard, T. Cytology of the life-cycle of Morchella. Mycol. Res. 1990, 94, 399–406. [Google Scholar] [CrossRef]
- Karakehian, J.M.; Quijada, L.; Pfister, D.H.; Tocci, G.E.; Miller, A.N. Methods for observing, culturing, and studying living ascospores. Asian J. Mycol. 2021, 4, 1–18. [Google Scholar]
- Richard, F.; Bellanger, J.-M.; Clowez, P.; Hansen, K.; O’donnell, K.; Urban, A.; Sauve, M.; Courtecuisse, R.; Moreau, P.-A. True morels (Morchella, Pezizales) of Europe and North America: Evolutionary relationships inferred from multilocus data and a unified taxonomy. Mycologia 2015, 107, 359–382. [Google Scholar] [CrossRef]
- Seaver, J. The North American Cup-Fungi (Operculates) Supplemented Edition; Published by the author: New York, NY, USA, 1942. [Google Scholar]
- van Vooren, N.; Dougoud, R.; Moyne, G.; Vega, M.; Carbone, M.; Perić, B. Overview of violet Peziza (Pezizales) present in Europe. 2nd part: The genus Phylloscypha. Ascomycete.org 2021, 13, 107–116. [Google Scholar]
- Marx, D.H. The influence of ectotrophic mycorrhizal fungi on the resistance of pine roots to pathogenic infection, I. Antagonism of mycorrhizal fungi to root pathogenic fungi and soil bacteria. Phytopathology 1969, 59, 153–163. [Google Scholar]
- Hunter, J.E.; Steadman, J.R.; Cigna, J.A. Preservation of ascospores of Sclerotinia sclerotiorum on membrane filters. Phytopathology 1982, 72, 650–652. [Google Scholar] [CrossRef]
- Gomez-Lopez, A.; Aberkane, A.; Petrikkou, E.; Mellado, E.; Rodriguez-Tudela, J.L.; Cuenca-Estrella, M. Analysis of the influence of Tween concentration, inoculum size, assay medium, and reading time on susceptibility testing of Aspergillus spp. J. Clin. Microbiol. 2005, 43, 1251–1255. [Google Scholar] [CrossRef] [PubMed]
- Castellani, A. Long term observations on pathogenic fungi in culture. J. Trop. Med. Hyg. 1961, 64, 60–63. [Google Scholar]
- Karabıçak, N.; Karatuna, O.; Akyar, I. Evaluation of the Viabilities and Stabilities of Pathogenic Mold and Yeast Species Using Three Different Preservation Methods Over a 12-Year Period Along with a Review of Published Reports. Mycopathologia 2016, 181, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Vandepol, N.; Liber, J.; Desirò, A.; Na, H.; Kennedy, M.; Barry, K.; Grigoriev, I.V.; Miller, A.N.; O’Donnell, K.; Stajich, J.E.; et al. Resolving the Mortierellaceae phylogeny through synthesis of multi-gene phylogenetics and phylogenomics. Fungal Divers. 2020, 104, 267–289. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Fungal Barcoding Consortium; Fungal Barcoding Consortium Author List; Bolchacova, E.; et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Katoh, K.; Toh, H. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 2010, 26, 1899–1900. [Google Scholar] [CrossRef]
- Rambaut, A. Se-Al: Sequence Alignment Editor, Version 2.0a11. 2007. Available online: http://tree.bio.ed.ac.uk/software/seal/ (accessed on 1 March 2023).
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Dirks, A.C.; Mohamed, O.G.; Schultz, P.J.; Miller, A.N.; Tripathi, A.; James, T.Y. Not all bad: Gyromitrin has a limited distribution in the false morels as determined by a new ultra high-performance liquid chromatography method. Mycologia 2023, 115, 1–15. [Google Scholar] [CrossRef]
- Romero, D.D.; Rivera, H.V.; Reyes, B.R.; Reyes, J.A.; Campos, A.R.M. Isolation and purification of ectomycorrhizal fungus Helvella lacunosa in different culture media. Trop. Subtrop. Agrosystems 2013, 16, 51–59. [Google Scholar]
- Iotti, M.; Piattoni, F.; Zambonelli, A. Techniques for host plant inoculation with truffles and other edible ectomycorrhizal mushrooms. In Edible Ectomycorrhizal Mushrooms: Current Knowledge and Future Prospects; Springer: Berlin/Heidelberg, Germany, 2013; pp. 145–161. [Google Scholar]
- Lorrain, C.; Gonçalves dos Santos, K.C.; Germain, H.; Hecker, A.; Duplessis, S. Advances in understanding obligate biotrophy in rust fungi. New Phytol. 2019, 222, 1190–1206. [Google Scholar] [CrossRef]
- Luginbuehl, L.H.; Menard, G.N.; Kurup, S.; Van Erp, H.; Radhakrishnan, G.V.; Breakspear, A.; Oldroyd, G.E.D.; Eastmond, P.J. Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 2017, 356, 1175–1178. [Google Scholar] [CrossRef]
- Ogawa, S.; Nagao, H.; Ando, A.; Nagata, Y. Enhancement of ascospore germination from Aleuria aurantia after cold storage. Mycoscience 2000, 41, 287–289. [Google Scholar] [CrossRef]
- Fries, N. Untersuchungen über Sporenkeimung und Mycelentwicklung bodenbewohnender Hymenomyceten; Acta Universitatis Upsaliensis: Uppsala, Sweden, 1943; Volume 6, pp. 1–81. [Google Scholar]
- Fries, N. Spore germination in Boletus induced by amino acids. Proc. K. Ned. Akad. Wet. Ser. C 1976, 79, 142–146. [Google Scholar]
- Fries, N. Basidiospore germination in some mycorrhiza-forming Hymenomycetes;Trans. Br. Mycol. Soc. 1978, 70, 319–324. [Google Scholar] [CrossRef]
- Miller, S.L.; Torres, P.; McClean, T.M. Basidiospore viability and germination in ectomycorrhizal and saprotrophic basidiomycetes. Mycol. Res. 1993, 97, 141–149. [Google Scholar] [CrossRef]
- Bohorquez, J.; Nilsen, A.; Larcombe, M.; Orlovich, D.; Lord, J. Spore viability and germination of some ectomycorrhizal fungi from New Zealand and implications for forest restoration. N. Z. J. Bot. 2021, 59, 250–266. [Google Scholar] [CrossRef]
- Ishida, T.A.; Nara, K.; Tanaka, M.; Kinoshita, A.; Hogetsu, T. Germination and infectivity of ectomycorrhizal fungal spores in relation to their ecological traits during primary succession. New Phytol. 2008, 180, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Norman, J.E.; Egger, K.N. Phylogeny of the genus Plicaria and its relationship to Peziza inferred from ribosomal DNA sequence analysis. Mycologia 1996, 88, 986–995. [Google Scholar] [CrossRef]
- Tedersoo, L.; May, T.W.; Smith, M.E. Ectomycorrhizal lifestyle in fungi: Global diversity, distribution, and evolution of phylogenetic lineages. Mycorrhiza. 2010, 20, 217–263. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Cai, Y.; He, P.; Zhang, Y.; Bian, Y. Ascospore germination and nuclear behavior of Morchella crassipes. Acta Edulis Fungi 2017, 24, 18–20. [Google Scholar]
- Paden, J.W. The Conidial State of Peziza brunneoatra. Mycologia 1967, 59, 932–934. [Google Scholar] [CrossRef]
- Paden, J.W. Ascospore germination, growth in culture, and imperfect spore formation in Cookeina sulcipes and Phillipsia crispata. Canad. J. Bot. 1975, 53, 56–61. [Google Scholar] [CrossRef]
- Öpik, M.; Kullman, B.; Kollom, A. Sarcoscypha austriaca (Pezizales) in Estonia. Folia Cryptogamica Estonica 2000, 36, 107–112. [Google Scholar]
- Boedijn, K.B. The genera Phillipsia and Cookeina in Netherlands India. Bull. Jard. Bot. Buitenzorg. Serie III 1933, 13, 57–76. [Google Scholar]
- Alvarado-Castillo, G.; Mata, G.; Pérez-Vázquez, A.; Martínez-Carrera, D.; Nava-Tablada, M.E.; Gallardo-López, F.; Osorio-Acosta, F. Sclerotia formation in Morchella esculenta and M. conica in vitro. Rev. Mex. Mic. 2012, 35, 35–41. [Google Scholar]
- Yuan, B.H.; Li, H.; Liu, L.; Du, X.H. Successful induction and recognition of conidiation, conidial germination and chlamydospore formation in pure culture of Morchella. Fungal Biol. 2021, 125, 285–293. [Google Scholar] [CrossRef]
- Aver’yanov, A.A.; Lapikova, V.P.; Pasechnik, T.D.; Zakharenkova, T.S.; Baker, C.J. Self-inhibition of spore germination via reactive oxygen in the fungus Cladosporium cucumerinum, causal agent of cucurbit scab. Eur. J. Plant Pathol. 2011, 130, 541–550. [Google Scholar] [CrossRef]
- Baral Hans-Otto, A. Monograph of Helicogonium (=Myriogonium, Leotiales), a Group of Non-Ascocarpous Intrahymenial Mycoparasites. Nova Hedwigia 1999, 69, 1–71. [Google Scholar] [CrossRef]


| Herbarium Number | Taxon | Collection Date | Date of Inoculation | Days until Germination | GenBank Number |
|---|---|---|---|---|---|
| FLAS-F-68218 | Galiella rufa | 16 August 2021 | 14 February 2022 | 5 | OM672689 |
| FLAS-F-68574 | Galiella rufa | 12 August 2021 | 17 January 2022 | 7 | OM672856 |
| MSC0286084 | Gyromitra venenata | 23 May 2021 | 31 March 2022 | 1 | OQ413215 |
| MSC0286085 | Morchella angusticeps | 1 May 2021 | 5 April 2022 | 1 | OQ413216 |
| FLAS-F-68233 | Peziza griseorosea | 16 August 2021 | 13 January 2022 | 11 | OM672695 |
| FLAS-F-68375 | Peziza griseorosea | 13 August 2021 | 13 January 2022 | 1 | OM672751 |
| MSC0276205 | Peziza sp. 2 | 31 March 2021 | 28 March 2022 | 26 | OQ413218 |
| MSC0286082 | Peziza varia complex sp. | 23 May 2021 | 12 April 2022 | 2 | OQ413219 |
| MSC0286086 | Peziza varia complex sp. | 15 October 2021 | 25 January 2022 | 3 | OQ413220 |
| MSC0286088 | Peziza varia complex sp. | 11 October 2021 | 21 February 2022 | 1 | OQ413221 |
| MSC0286087 | Peziza varia complex sp. | 17 April 2021 | 28 March 2022 | 1 | OQ413222 |
| MSC0286080 | Peziza sp. 1 | 5 October 2020 | 31 March 2022 | 12 | OQ413223 |
| MSC0286083 | Phylloscypha phyllogena | 23 May 2021 | 12 April 2022 | 13 | OQ413217 |
| MSC0286081 | Pseudoplectania sp. | 23 May 2021 | 28 March 2022 | 1 | OQ413224 |
| FLAS-F-68215 | Sarcoscypha occidentalis | 16 August 2021 | 31 January 2022 | 8 | OM672687 |
| FLAS-F-68662 | Sarcoscypha occidentalis | 8 September 2021 | 14 February 2022 | 9 | OM672899 |
| FLAS-F-68510 | Scutellinia sp. 2 | 7 September 2021 | 31 January 2022 | 15 | OM672830 |
| MSC0286078 | Scutellinia sp. 1 | 12 July 2021 | 5 April 2022 | 10 | OQ413225 |
| JV358 | Sphaerosporella brunnea | 6 October 2021 | 18 February 2022 | 5 | OQ413226 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sow, A.; Van Wyk, J.; Lemmond, B.; Healy, R.; Smith, M.E.; Bonito, G. Effective Field Collection of Pezizales Ascospores for Procuring Diverse Fungal Isolates. Diversity 2024, 16, 165. https://doi.org/10.3390/d16030165
Sow A, Van Wyk J, Lemmond B, Healy R, Smith ME, Bonito G. Effective Field Collection of Pezizales Ascospores for Procuring Diverse Fungal Isolates. Diversity. 2024; 16(3):165. https://doi.org/10.3390/d16030165
Chicago/Turabian StyleSow, Alassane, Judson Van Wyk, Benjamin Lemmond, Rosanne Healy, Matthew E. Smith, and Gregory Bonito. 2024. "Effective Field Collection of Pezizales Ascospores for Procuring Diverse Fungal Isolates" Diversity 16, no. 3: 165. https://doi.org/10.3390/d16030165
APA StyleSow, A., Van Wyk, J., Lemmond, B., Healy, R., Smith, M. E., & Bonito, G. (2024). Effective Field Collection of Pezizales Ascospores for Procuring Diverse Fungal Isolates. Diversity, 16(3), 165. https://doi.org/10.3390/d16030165






