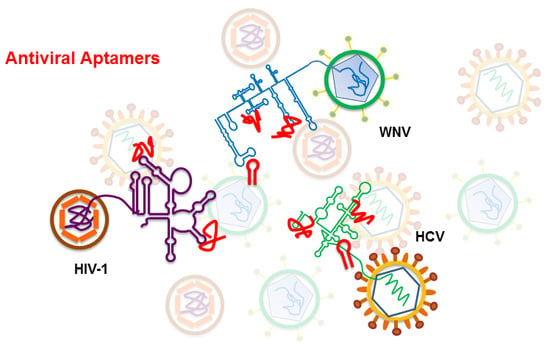
Two Examples of RNA Aptamers with Antiviral Activity. Are Aptamers the Wished Antiviral Drugs?
Abstract
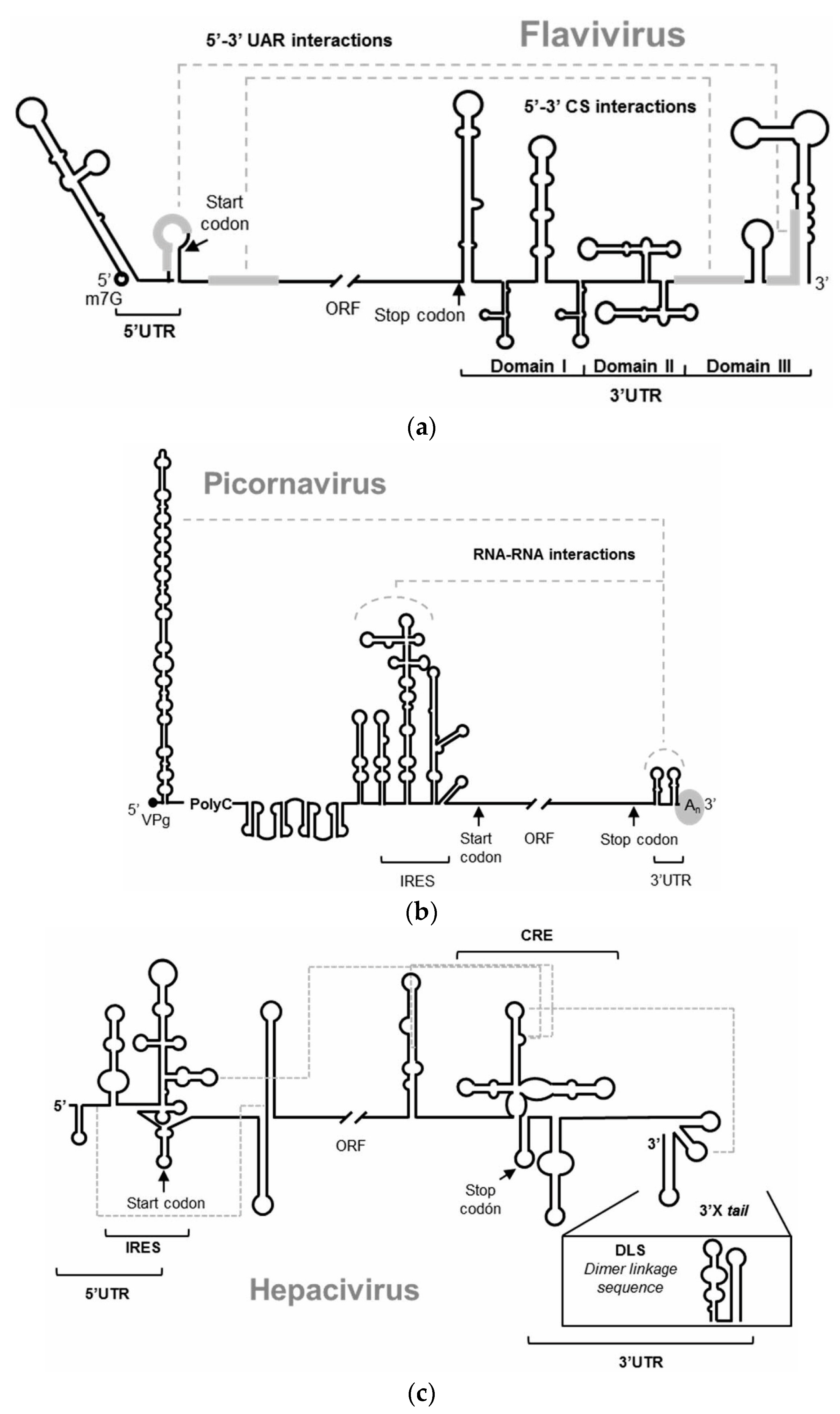
:1. Introduction
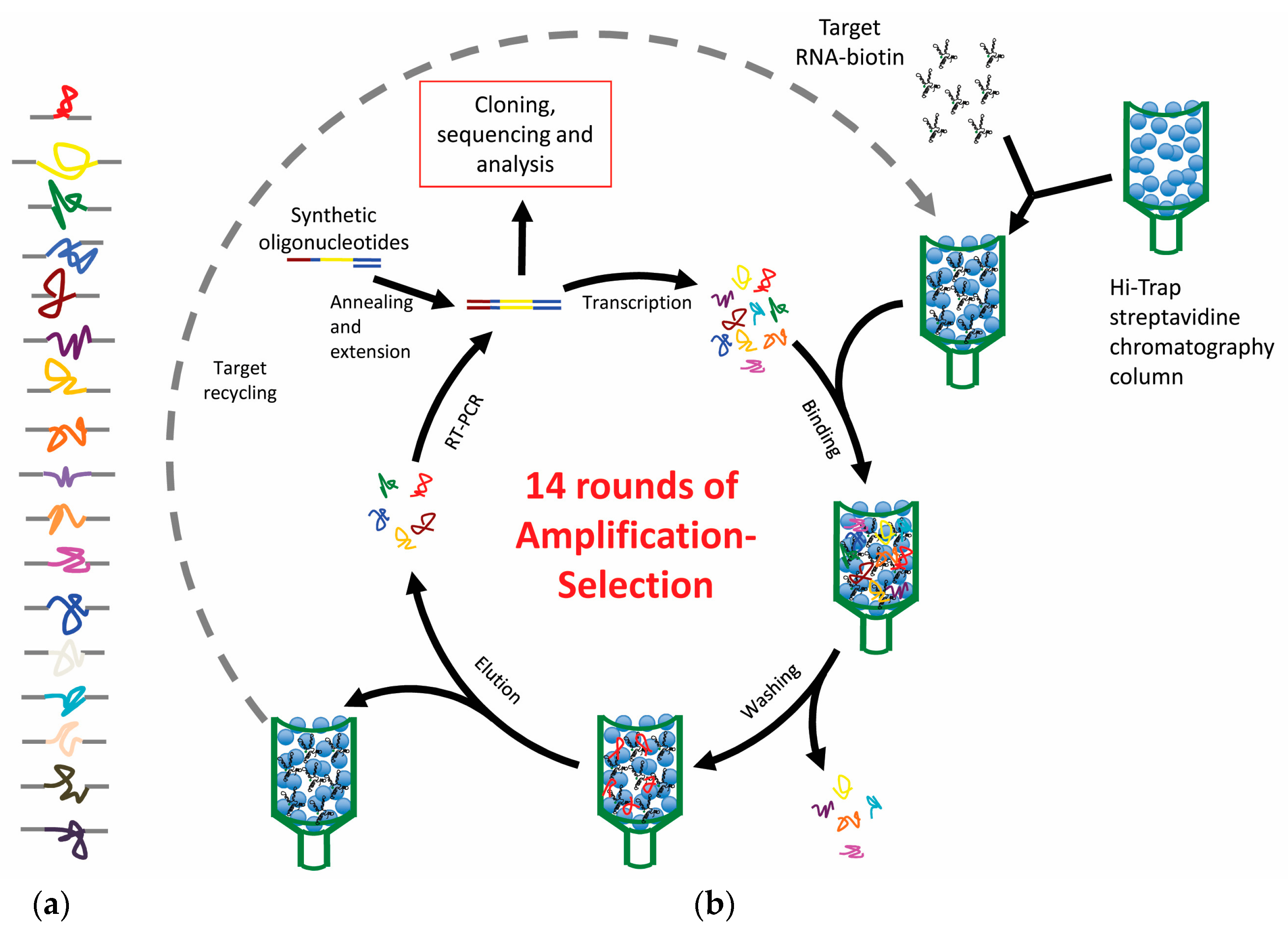
2. Aptamers, Selection Procedure, and Features
2.1. SELEX
2.2. Aptamer Features
3. Antiviral Aptamers
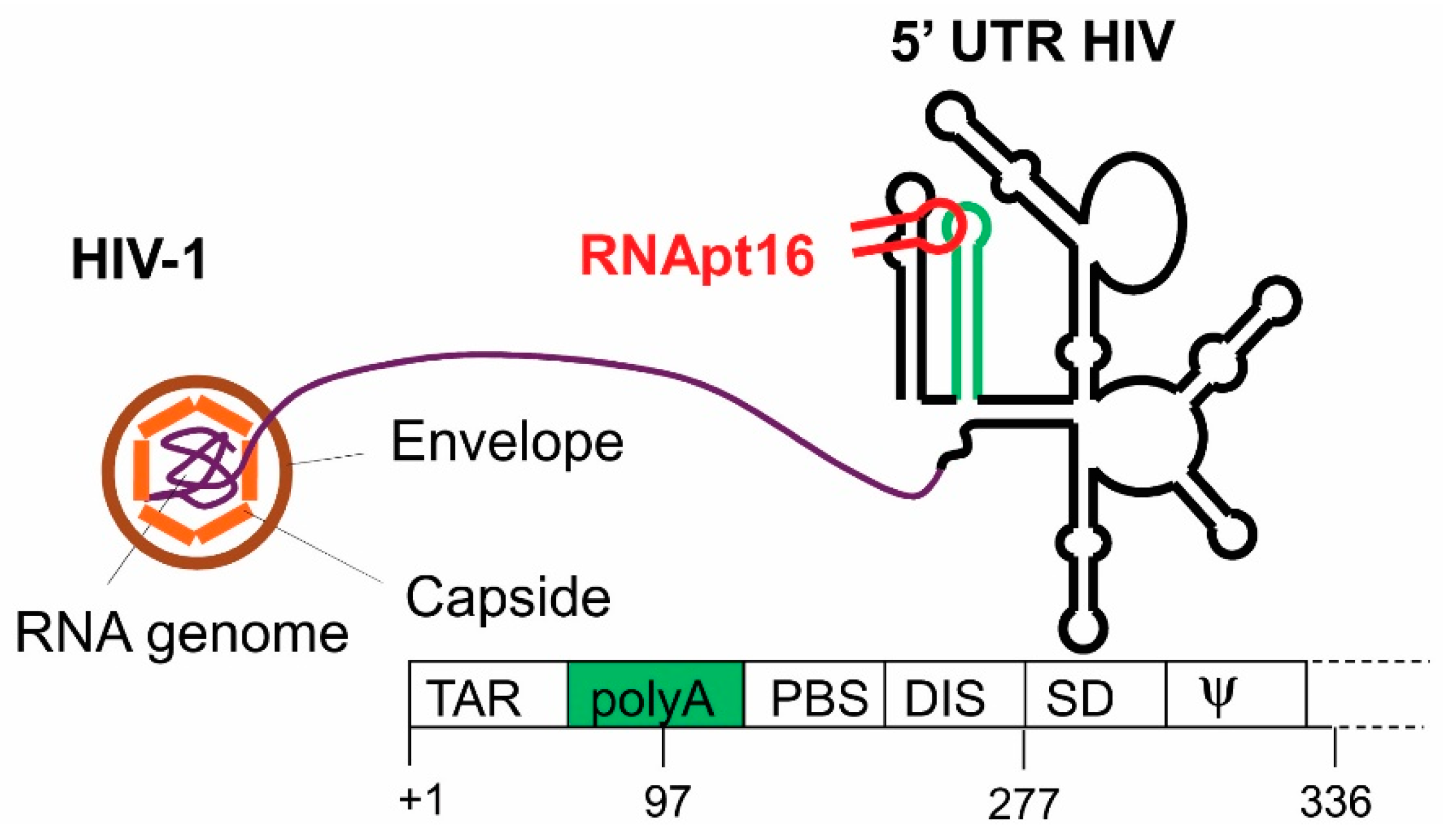
3.1. Anti HIV-1 RNA Aptamers
3.2. Anti HCV RNA Aptamers
4. Conclusions
Funding
Conflicts of Interest
References
- Romero-López, C.; Berzal-Herranz, A. Unmasking the information encoded as structural motifs of viral RNA genomes: A potential antiviral target. Rev. Med. Virol. 2013, 23, 340–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moomau, C.; Musalgaonkar, S.; Khan, Y.A.; Jones, J.E.; Dinman, J.D. Structural and functional characterization of programmed ribosomal frameshift signals in west nile virus strains reveals high structural plasticity among cis-acting RNA elements. J. Biol. Chem. 2016, 291, 15788–15795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kendra, J.A.; Advani, V.M.; Chen, B.; Briggs, J.W.; Zhu, J.; Bress, H.J.; Pathy, S.M.; Dinman, J.D. Functional and structural characterization of the chikungunya virus translational recoding signals. J. Biol. Chem. 2018, 293, 17536–17545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature 1992, 355, 850–852. [Google Scholar] [CrossRef]
- Schneider, D.; Tuerk, C.; Gold, L. Selection of high affinity RNA ligands to the bacteriophage R17 coat protein. J. Mol. Biol. 1992, 228, 862–869. [Google Scholar] [CrossRef]
- Wilson, D.S.; Szostak, J.W. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999, 68, 611–647. [Google Scholar] [CrossRef] [Green Version]
- Rajendran, M.; Ellington, A.D. Selection of fluorescent aptamer beacons that light up in the presence of zinc. Anal. Bioanal. Chem. 2008, 390, 1067–1075. [Google Scholar] [CrossRef]
- Raddatz, M.S.; Dolf, A.; Endl, E.; Knolle, P.; Famulok, M.; Mayer, G. Enrichment of cell-targeting and population-specific aptamers by fluorescence-activated cell sorting. Angew. Chem. Int. Ed. Engl. 2008, 47, 5190–5193. [Google Scholar] [CrossRef]
- Torres-Chavolla, E.; Alocilja, E.C. Aptasensors for detection of microbial and viral pathogens. Biosens. Bioelectron. 2009, 24, 3175–3182. [Google Scholar] [CrossRef] [PubMed]
- Marton, S.; Reyes-Darias, J.A.; Sánchez-Luque, F.J.; Romero-Lopez, C.; Berzal-Herranz, A. In vitro and ex vivo selection procedures for identifying potentially therapeutic DNA and RNA molecules. Molecules 2010, 15, 4610–4638. [Google Scholar] [CrossRef] [Green Version]
- Ku, T.H.; Zhang, T.; Luo, H.; Yen, T.M.; Chen, P.W.; Han, Y.; Lo, Y.H. Nucleic acid aptamers: An emerging tool for biotechnology and biomedical sensing. Sensors 2015, 15, 16281–16313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, P.K. Monitoring intact viruses using aptamers. Biosensors 2016, 6, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng, E.W.; Adamis, A.P. Anti-VEGF aptamer (pegaptanib) therapy for ocular vascular diseases. Ann. N. Y. Acad. Sci. 2006, 1082, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Ng, E.W.; Shima, D.T.; Calias, P.; Cunningham, E.T., Jr.; Guyer, D.R.; Adamis, A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006, 5, 123–132. [Google Scholar] [CrossRef]
- Ciulla, T.A.; Rosenfeld, P.J. Antivascular endothelial growth factor therapy for neovascular age-related macular degeneration. Curr. Opin. Ophthalmol. 2009, 20, 158–165. [Google Scholar] [CrossRef]
- Gopinath, S.C. Methods developed for SELEX. Anal. Bioanal. Chem. 2007, 387, 171–182. [Google Scholar] [CrossRef]
- Tombelli, S.; Minunni, M.; Mascini, M. Analytical applications of aptamers. Biosens. Bioelectron. 2005, 20, 2424–2434. [Google Scholar] [CrossRef]
- Odeh, F.; Nsairat, H.; Alshaer, W.; Ismail, M.A.; Esawi, E.; Qaqish, B.; Bawab, A.A.; Ismail, S.I. Aptamers chemistry: Chemical modifications and conjugation strategies. Molecules 2019, 25, 3. [Google Scholar] [CrossRef] [Green Version]
- Berzal-Herranz, A.; Romero-Lopez, C. RNA Aptamers: Antiviral Drugs of the Future. In Proceedings of the 5th International Electronic Conference in Medicinal Chemistry, MDPI AG: Sciforum. 12 November 2019. Available online: https://ecmc2019.sciforum.net/ (accessed on 22 July 2020). [CrossRef] [Green Version]
- Theissen, G.; Richter, A.; Lukacs, N. Degree of biotinylation in nucleic acids estimated by a gel retardation assay. Anal. Biochem. 1989, 179, 98–105. [Google Scholar] [CrossRef]
- Romero-López, C.; Barroso-delJesus, A.; Puerta-Fernández, E.; Berzal-Herranz, A. Interfering with hepatitis C virus IRES activity using RNA molecules identified by a novel in vitro selection method. Biol. Chem. 2005, 386, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Marton, S.; Berzal-Herranz, B.; Garmendia, E.; Cueto, F.J.; Berzal-Herranz, A. Anti-HCV RNA aptamers targeting the genomic cis-acting replication element. Pharmaceuticals 2012, 5, 49–60. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Luque, F.J.; Stich, M.; Manrubia, S.; Briones, C.; Berzal-Herranz, A. Efficient HIV-1 inhibition by a 16 nt-long RNA aptamer designed by combining in vitro selection and in silico optimisation strategies. Sci. Rep. 2014, 4, 6242. [Google Scholar] [CrossRef] [PubMed]
- Berkhout, B. HIV-1 as RNA evolution machine. RNA Biol. 2011, 8, 225–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berzal-Herranz, A.; Romero-López, C.; Berzal-Herranz, B.; Ramos-Lorente, S. Potential of the other genetic information coded by the viral RNA genomes as antiviral target. Pharmaceuticals 2019, 12, 38. [Google Scholar] [CrossRef] [Green Version]
- Romero-López, C.; Berzal-Herranz, A. The role of the RNA-RNA interactome in the hepatitis C virus life cycle. Int. J. Mol. Sci. 2020, 21, 1479. [Google Scholar] [CrossRef] [Green Version]
- Romero-López, C.; Berzal-Herranz, A. The 5BSL3.2 functional RNA domain connects distant regions in the hepatitis C virus genome. Front. Microbiol. 2017, 8, 2093. [Google Scholar] [CrossRef]
- Fernández-Sanlés, A.; Berzal-Herranz, B.; González-Matamala, R.; Ríos-Marco, P.; Romero-López, C.; Berzal-Herranz, A. RNA aptamers as molecular tools to study the functionality of the hepatitis C virus CRE region. Molecules 2015, 20, 16030–16047. [Google Scholar] [CrossRef]
- Marton, S.; Romero-López, C.; Berzal-Herranz, A. RNA aptamer-mediated interference of HCV replication by targeting the CRE-5BSL3.2 domain. J. Viral Hepat. 2013, 20, 103–112. [Google Scholar] [CrossRef] [Green Version]
- Romero-López, C.; Díaz-González, R.; Berzal-Herranz, A. Inhibition of hepatitis C virus internal ribosome entry site-mediated translation by an RNA targeting the conserved IIIf domain. Cell Mol. Life Sci. 2007, 64, 2994–3006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero-López, C.; Díaz-González, R.; Barroso-delJesus, A.; Berzal-Herranz, A. Inhibition of hepatitis C virus replication and internal ribosome entry site-dependent translation by an RNA molecule. J. Gen. Virol. 2009, 90, 1659–1669. [Google Scholar] [CrossRef] [PubMed]
- Romero-López, C.; Berzal-Herranz, B.; Gómez, J.; Berzal-Herranz, A. An engineered inhibitor RNA that efficiently interferes with hepatitis C virus translation and replication. Antivir. Res. 2012, 94, 131–138. [Google Scholar] [CrossRef] [Green Version]
- Romero-López, C.; Lahlali, T.; Berzal-Herranz, B.; Berzal-Herranz, A. Development of optimized inhibitor RNAs allowing multisite-targeting of the HCV genome. Molecules 2017, 22, 861. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Nº Repetitions | Aptamer 1 |
|---|---|
| 23 | CACCACUAUUGUUGGCAAGGAAGCA |
| 6 | GUACGGCAAGGAGUACAUCGUAGCA |
| 2 | CACAACCUGGGUGGCAAGGAACCCA |
| 1 | CACCGCUAUUGUUGGCAAGGAAGCA |
| 1 | CACCACUAUUGUUGGCAAGGAAGCA |
| 1 | GUACGGCAAGGAGUACAUCGCAGCA |
| 1 | CACUACUCUACGGCUCGAAGCCCCA |
| 1 | AACCACAACGGCUAACCACUGCCCA |
| 1 | CACUACCGACCGUCCACACCAGCCA |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berzal-Herranz, A.; Romero-López, C. Two Examples of RNA Aptamers with Antiviral Activity. Are Aptamers the Wished Antiviral Drugs? Pharmaceuticals 2020, 13, 157. https://doi.org/10.3390/ph13080157
Berzal-Herranz A, Romero-López C. Two Examples of RNA Aptamers with Antiviral Activity. Are Aptamers the Wished Antiviral Drugs? Pharmaceuticals. 2020; 13(8):157. https://doi.org/10.3390/ph13080157
Chicago/Turabian StyleBerzal-Herranz, Alfredo, and Cristina Romero-López. 2020. "Two Examples of RNA Aptamers with Antiviral Activity. Are Aptamers the Wished Antiviral Drugs?" Pharmaceuticals 13, no. 8: 157. https://doi.org/10.3390/ph13080157
APA StyleBerzal-Herranz, A., & Romero-López, C. (2020). Two Examples of RNA Aptamers with Antiviral Activity. Are Aptamers the Wished Antiviral Drugs? Pharmaceuticals, 13(8), 157. https://doi.org/10.3390/ph13080157