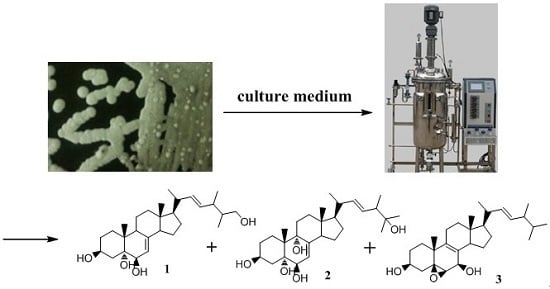
Ergosterols from the Culture Broth of Marine Streptomyces anandii H41-59
Abstract
:1. Introduction
2. Results and Discussion
2.1. Characterization of the Compounds
2.2. Biological Activity
3. Experimental Section
3.1. General Experimental Procedures
3.2. Strain Isolation and Identification
3.3. Fermentation and Extraction
3.4. Isolation and Purification
3.5. Biological Activities
3.5.1. Antimicrobial Activity
3.5.2. Cytotoxicity Assay
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| HPLC | High performance liquid chromatography |
| DEPT | Distortionless enhancement by polarization transfer |
| COSY | Two dimensional 1H correlation |
| HSQC | 1H-detected heteronuclear single-quantum coherence |
| HMBC | 1H-detected heteronuclear multiple-bond correlation |
| NOESY | Nuclear overhauser effect spectroscopy |
| MTT | 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide |
References
- Podar, M.; Reysenbach, A.L. New opportunities revealed by biotechnological explorations of extremophiles. Curr. Opin. Biotechnol. 2006, 17, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Saikia, S.; Kolita, B.; Dutta, P.P.; Dutta, D.J.; Neipihoi; Nath, S.; Bordoloi, M.; Quan, P.M.; Thuy, T.T.; Phuong, D.L.; et al. Marine steroids as potential anticancer drug candidates: In silico investigation in search of inhibitors of Bcl-2 and CDK-4/Cyclin D1. Steroids 2015, 102, 7–16. [Google Scholar] [PubMed]
- Han, L.D.; Cui, J.G.; Huang, C.S. Bioactive polyhydroxy sterols and sapogenins from marine organisms. Chin. J. Org. Chem. 2003, 3, 305–311. [Google Scholar]
- Tremarin, A.; Longhi, D.A.; Salomão, B.D.; Aragão, G.M. Modeling the growth of Byssochlamys fulva and Neosartorya fischeri onsolidified apple juice by measuring colony diameter andergosterol content. Int. J. Food Microbiol. 2015, 193, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Iaea, D.B.; Maxfield, F.R. Cholesterol trafficking and distribution. Essays Biochem. 2015, 57, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2015, 32, 116–211. [Google Scholar] [CrossRef] [PubMed]
- Rateb, M.E.; Hallyburton, I.; Houssen, W.E.; Bull, A.T.; Goodfellow, M.; Rakesh, S.; Jaspars, M.; Ebel, R. Induction of diverse secondary metabolites in Aspergillus fumigatus by microbial co-culture. RSC Adv. 2013, 3, 14444–14450. [Google Scholar] [CrossRef] [Green Version]
- Toumatia, O.; Yekkour, A.; Goudjal, Y.; Riba, A.; Coppel, Y.; Mathieu, F.; Sabaou, N.; Zitouni, A. Antifungal properties of an actinomycin D-producing strain, Streptomyces sp. IA1, isolated from a Saharan soil. J. Basic Microbiol. 2015, 55, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Triton, T.R.; Yee, G. The anticancer agent adriamycin can be actively cytotoxic without entering cells. Science 1982, 217, 248–250. [Google Scholar] [CrossRef] [PubMed]
- Kino, T.; Hatanaka, H.; Miyata, S.; Inamura, N.; Nishiyama, M.; Yajima, T.; Goto, T.; Okuhara, M.; Kohsaka, M.; Aoki, H. FK-506, a novel immunosuppressant isolated from a Streptomyces. II. Immunosuppressive effect of FK-506 in vitro. J. Antibiot. 1987, 40, 1256–1265. [Google Scholar] [CrossRef] [PubMed]
- Piccialli, V.; Sica, D. Four new trihydroxylated sterols from the sponge Spongionella gracilis. J. Nat. Prod. 1987, 50, 915–920. [Google Scholar] [CrossRef]
- Wang, X.N.; Du, J.C.; Tan, R.X.; Liu, J.K. Chemical constituents of basidiomycete Hydnum repandum. Chin. Tradit. Herb Drugs 2005, 36, 1126–1130. [Google Scholar]
- Qi, S.H.; Zhang, S.; Wang, Y.F.; Li, M.Y. Complete 1H and 13C NMR assignments of three new polyhydroxylated sterols from the South China Sea gorgonian Subergorgia suberosa. Magn. Reson. Chem. 2007, 45, 1088–1091. [Google Scholar] [CrossRef] [PubMed]
- Kawagishi, H.; Katsumi, R.; Sazaw, T.; Mizuno, T.; Hagiwara, T.; Nakamura, T. Cytotoxic steroids from the mushroom Agaricus blazei. Phytochemistry 1988, 27, 2777–2779. [Google Scholar] [CrossRef]
- Hu, X.L.; Xu, W.F.; Lu, X.; Wu, X.; Bai, J.; Pei, Y.H. Secondary metabolites from endophyte fungus Fusariums sp. LC-1. Chin. Pharm. J. 2013, 48, 17–21. [Google Scholar]
- Kobayashi, M.; Kanda, F. Marine sterols. 18. Isolation and structure of four novel oxygenated sterols from a Gorgonian coral Melithaea ocracea. J. Chem. Soc. Perkin Trans. 1 1991, 22, 1177–1179. [Google Scholar] [CrossRef]
- Huang, X.C.; Liu, H.L.; Guo, Y.W. Chemical constituents of marine sponge Biemna fortis Topsent. Chin. J. Nat. Med. 2008, 6, 348–353. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, S.J.; Mo, S.Y.; Li, S. An abietane diterpene and a sterol from fungus Phellinus igniarius. Chin. Chem. Lett. 2006, 17, 481–484. [Google Scholar]
- Xiong, H.Y.; Fei, D.Q.; Zhou, J.S.; Yang, C.J.; Ma, G.L. Steroids and other constituents from the mushroom Armillaria lueo-virens. Chem. Nat. Compounds 2009, 45, 759–762. [Google Scholar] [CrossRef]
- Li, H.J.; Lin, Y.C.; Vrijmoed, L.L.P. A new cytotoxic sterol produced by an endophytic fungus from Castaniopsis fissa at the South China Sea coast. Chin. Chem. Lett. 2004, 15, 419–422. [Google Scholar]
- Yi, S.; Li, T.; Jian, H.; Wen, L.; Yue, H.P. Cytotoxic sterols from marine-derived fungus Pennicillium sp. Nat. Prod. Research 2006, 20, 381–384. [Google Scholar]
- Noboru, S.; Hideyuki, T.; Kazuo, V. Sterol analysis of DMI-resistant and sensitive strains of Venturia inaequalis. Phytochemistry 1996, 41, 1301. [Google Scholar]
- Ishizuka, T.; Yaoita, Y.; Kikuchi, M. Sterol constituents from the fruit bodies of Grifola frondosa (FR.) S. F. Gray. Chem. Pharm. Bull 1997, 45, 1756–1760. [Google Scholar] [CrossRef]
- Ishizaki, T.; Hirayama, N.; Shinkawa, H.; Nimi, O.; Murooka, Y. Nucleotide sequence of the gene for cholesterol oxidase from a Streptomyces sp. J. Bacteriol. 1989, 171, 596–601. [Google Scholar] [PubMed]
- Mendes, M.V.; Recio, E.; Antón, N.; Guerra, S.M.; Santos-Aberturas, J.; Martin, J.F.; Aparicio, J.F. Cholesterol oxidases act as signaling proteins for the biosynthesis of the polyene macrolide pimaricin. Chem. Biol. 2007, 14, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Maclachlan, J.; Wotherspoon, A.T.; Ansell, R.O.; Brooks, C.J. Cholesterol oxidase: sources, physical properties and analytical applications. J. Steroid Biochem. Mol. Biol. 2000, 72, 169–195. [Google Scholar] [CrossRef]
- Nikolayeva, V.M.; Egorova, O.V.; Dovbnya, D.V.; Donova, M.V. Extracellular 3β-hydroxysteroid oxidase of Mycobacterium vaccae VKM Ac-1815D. J. Steroid Biochem. Mol. Biol. 2004, 91, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Drzyzga, O.; Fernández de Las, H.L.; Morales, V.; Navarro, L.J.M.; Perera, J. Cholesterol degradation by Gordonia cholesterolivorans. Appl. Environ. Microbiol. 2011, 77, 4802–4810. [Google Scholar] [CrossRef] [PubMed]
- Dovbnya, D.V.; Egorova, O.V.; Donova, M.V. Microbial side-chain degradation of ergosterol and its 3-substituted derivatives: A new route for obtaining of deltanoids. Steroids 2010, 75, 653–658. [Google Scholar] [CrossRef] [PubMed]
- Donova, M.V. Transformation of steroids by actinobacteria: A review. Appl. Biochem. Microbiol. 2007, 43, 1–14. [Google Scholar] [CrossRef]
- Paul, M.D. Medicinal Natural Products, 2nd ed.; Wiley: Hoboken, NJ, USA, 2005; pp. 235–236. ISBN 0470846275. [Google Scholar]
- Grdina, M.B.; Orfanopoulos, M.; Stephenson, L.M. A convenient synthetic sequence for the deuterium labeling of olefins in the allylic position. J. Org. Chem. 1979, 2936–2938. [Google Scholar] [CrossRef]
- Adam, W.; Griesbeck, A.; Staab, E. A convenient one-pot synthesis of epoxy alcohols via photooxygenation of olefins in the presence of titanium (IV) catalyst. Tetrahedron Lett. 1986, 27, 2839–2842. [Google Scholar] [CrossRef]





| No. | 1a | 2a | 3a | 3b |
|---|---|---|---|---|
| 1 | 1.30, m | 1.15, m | 1.61, m | 1.74, m |
| 1.47, m | 2.05, m | |||
| 2 | 1.24, m | 1.31, m | 1.45, m | 1.62, m |
| 1.60, m | 1.67, m | 1.75, m | 1.95, m | |
| 3 | 3.76, m | 3.76, m | 3.53, m | 3.91, m |
| 4 | 1.49, m | 1.47, m | 1.20, m | 1.45, m |
| 1.89, m | 1.91, m | 1.99, m | 2.20, m | |
| 5 | - | - | - | - |
| 6 | 3.37, m | 3.50, m | 2.87, d, 2.7 | 3.12, d, 2.7 |
| 7 | 5.08,m | 5.12, d, 4.5 | 4.10, brs | 4.36, d, 2.2 |
| 8 | - | - | - | - |
| 9 | 1.94, m | - | - | - |
| 10 | - | - | - | - |
| 11 | 1.24, m | 1.15, m | 1.97, m | 1.66, m |
| 1.66, m | 2.05, m | 2.07, m | ||
| 12 | 1.25, m | 1.51, m | 1.34, m | 1.45, m |
| 1.96, m | 1.68, m | 1.88, m | 1.95, m | |
| 13 | - | - | - | - |
| 14 | 1.80, m | 2.39, m | 2.00, m | 2.23, m |
| 15 | 1.36, m | 1.45, m | 1.23, m | 1.66, m |
| 1.48, m | 1.69, m | 2.07, m | ||
| 16 | 1.24, m | 1.25, m | 1.25, m | 1.30, m |
| 1.66, m | 1.68, m | 1.63, m | 1.80, m | |
| 17 | 1.25, m | 1.28, m | 1.14, m | 1.19, m |
| 18 | 0.54, s | 0.54, s | 0.57, s | 0.61, s |
| 19 | 0.91, s | 0.96, s | 1.17, s | 1.25, s |
| 20 | 2.00, m | 1.98, m | 2.00, m | 2.02, m |
| 21 | 0.99, d, 6.6 | 0.98, d, 6.1 | 0.98, d, 6.3 | 0.99, d, 6.6 |
| 22 | 5.19, dd, 15.4, 7.9 | 5.19, dd, 15.1, 7.8 | 5.18, dd, 15.1, 8.0 | 5.16, dd, 15.2, 7.6 |
| 23 | 5.21, dd, 15.4, 7.1 | 5.35, dd, 15.1, 7.0 | 5.20, dd, 15.1, 7.2 | 5.18, dd, 15.2, 6.9 |
| 24 | 2.15, m | 1.98, m | 1.84, m | 1.84, m |
| 25 | 1.41, m | 1.45, m | 1.43, m | |
| 26 | 0.74, d, 6.9 | 0.97, s | 0.79, d, 4.1 | 0.81, d, 4.5 |
| 27 | 3.16, m | 1.02, s | 0.80, d, 4.1 | 0.89, d, 4.5 |
| 3.32, m | ||||
| 28 | 0.92, d, 6.8 | 0.90, d, 6.8 | 0.88, d, 6.8 | 0.89, d, 6.8 |
| 3-OH | 4.21, d, 5.4 | 4.33, d, 3.6 | 4.68, brs | |
| 5-OH | 3.58, s | 5.00, s | ||
| 6-OH | 4.49, d, 5.6 | 4.76, d, 5.5 | ||
| 7-OH | 4.76, brs | |||
| 9-OH | 5.19, s | |||
| 25-OH | 4.08, s | |||
| 27-OH | 4.30, m |
| No. | 1a | 2a | 3a | 3b |
|---|---|---|---|---|
| 1 | 32.5 | 26.9 | 30.6 | 31.0 |
| 2 | 31.2 | 31.0 | 30.8 | 31.0 |
| 3 | 66.0 | 65.8 | 67.0 | 68.8 |
| 4 | 40.2 | 40.2 | 39.0 | 39.2 |
| 5 | 74.5 | 73.7 | 62.6 | 63.4 |
| 6 | 72.1 | 71.6 | 60.0 | 60.3 |
| 7 | 119.5 | 120.3 | 65.0 | 67.0 |
| 8 | 139.7 | 141.1 | 126.2 | 126.7 |
| 9 | 42.3 | 77.3 | 134.4 | 137.2 |
| 10 | 36.6 | 39.6 | 37.3 | 38.0 |
| 11 | 27.8 | 26.9 | 22.4 | 23.2 |
| 12 | 38.9 | 35.0 | 36.1 | 36.3 |
| 13 | 43.0 | 43.3 | 41.5 | 42.1 |
| 14 | 54.2 | 50.1 | 51.3 | 51.3 |
| 15 | 22.6 | 22.6 | 29.1 | 23.1 |
| 16 | 27.8 | 27.8 | 22.1 | 29.4 |
| 17 | 55.3 | 55.5 | 53.8 | 54.5 |
| 18 | 12.1 | 11.7 | 11.3 | 11.7 |
| 19 | 17.7 | 21.4 | 22.5 | 23.8 |
| 20 | 40.1 | 40.1 | 40.0 | 40.7 |
| 21 | 21.0 | 20.9 | 20.8 | 21.2 |
| 22 | 135.7 | 135.8 | 135.5 | 135.0 |
| 23 | 130.3 | 130.5 | 131.3 | 132.3 |
| 24 | 36.9 | 47.2 | 42.1 | 43.0 |
| 25 | 40.5 | 70.7 | 32.5 | 33.3 |
| 26 | 13.1 | 26.0 | 19.5 | 19.9 |
| 27 | 64.4 | 28.3 | 19.8 | 20.2 |
| 28 | 18.3 | 15.0 | 17.4 | 17.8 |
| Compounds | MCF-7 | SF-268 | NCI-H460 | Compounds | MCF-7 | SF-268 | NCI-H460 |
|---|---|---|---|---|---|---|---|
| 1 | >50 | >50 | >50 | 7 | >50 | >50 | >50 |
| 2 | >50 | >50 | >50 | 8 | 24.3 | 15.5 | 19.8 |
| 3 | 18.1 | 13.0 | 23.5 | 9 | >50 | >50 | >50 |
| 4 | >50 | >50 | >50 | 10 | 17.3 | 27.8 | 23.7 |
| 5 | >50 | >50 | >50 | 11 | 27.4 | 25.1 | 23.7 |
| 6 | >50 | >50 | >50 | 12 | >50 | >50 | >50 |
| Positive control | 4.0 | 41.0 | 25.1 | 13 | >50 | >50 | >50 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.-M.; Li, H.-Y.; Hu, C.; Sheng, H.-F.; Zhang, Y.; Lin, B.-R.; Zhou, G.-X. Ergosterols from the Culture Broth of Marine Streptomyces anandii H41-59. Mar. Drugs 2016, 14, 84. https://doi.org/10.3390/md14050084
Zhang Y-M, Li H-Y, Hu C, Sheng H-F, Zhang Y, Lin B-R, Zhou G-X. Ergosterols from the Culture Broth of Marine Streptomyces anandii H41-59. Marine Drugs. 2016; 14(5):84. https://doi.org/10.3390/md14050084
Chicago/Turabian StyleZhang, Yang-Mei, Hong-Yu Li, Chen Hu, Hui-Fan Sheng, Ying Zhang, Bi-Run Lin, and Guang-Xiong Zhou. 2016. "Ergosterols from the Culture Broth of Marine Streptomyces anandii H41-59" Marine Drugs 14, no. 5: 84. https://doi.org/10.3390/md14050084
APA StyleZhang, Y. -M., Li, H. -Y., Hu, C., Sheng, H. -F., Zhang, Y., Lin, B. -R., & Zhou, G. -X. (2016). Ergosterols from the Culture Broth of Marine Streptomyces anandii H41-59. Marine Drugs, 14(5), 84. https://doi.org/10.3390/md14050084