Molecular Cloning and Characterization of a Serotonin N-Acetyltransferase Gene, xoSNAT3, from Xanthomonas oryzae pv. oryzae
Abstract
:1. Introduction
2. Materials and Methods
2.1. General Information and Bacterial Strain
2.2. Identification of MT in Xoo
2.3. Phylogenetic Analysis of xoSNAT3 from Xoo Strain PXO99
2.4. Cloning of xoSNAT3 from Xoo Strain PXO99
2.5. Subcellular Localization of xoSNAT3 in Tobacco Leaves
2.6. Measurement of xoSNAT3 Enzymatic Activity In Vitro
2.7. Generation of xoSNAT3 Deletion Mutant
2.8. Complementation of the xoSNAT3 Mutant
2.9. RNA Extraction and Quantitative RT-PCR Analysis
2.10. Analysis of Pathogenicity and Biofilm Formation
2.11. Bioinformatics Analysis of xoSNAT3
3. Results
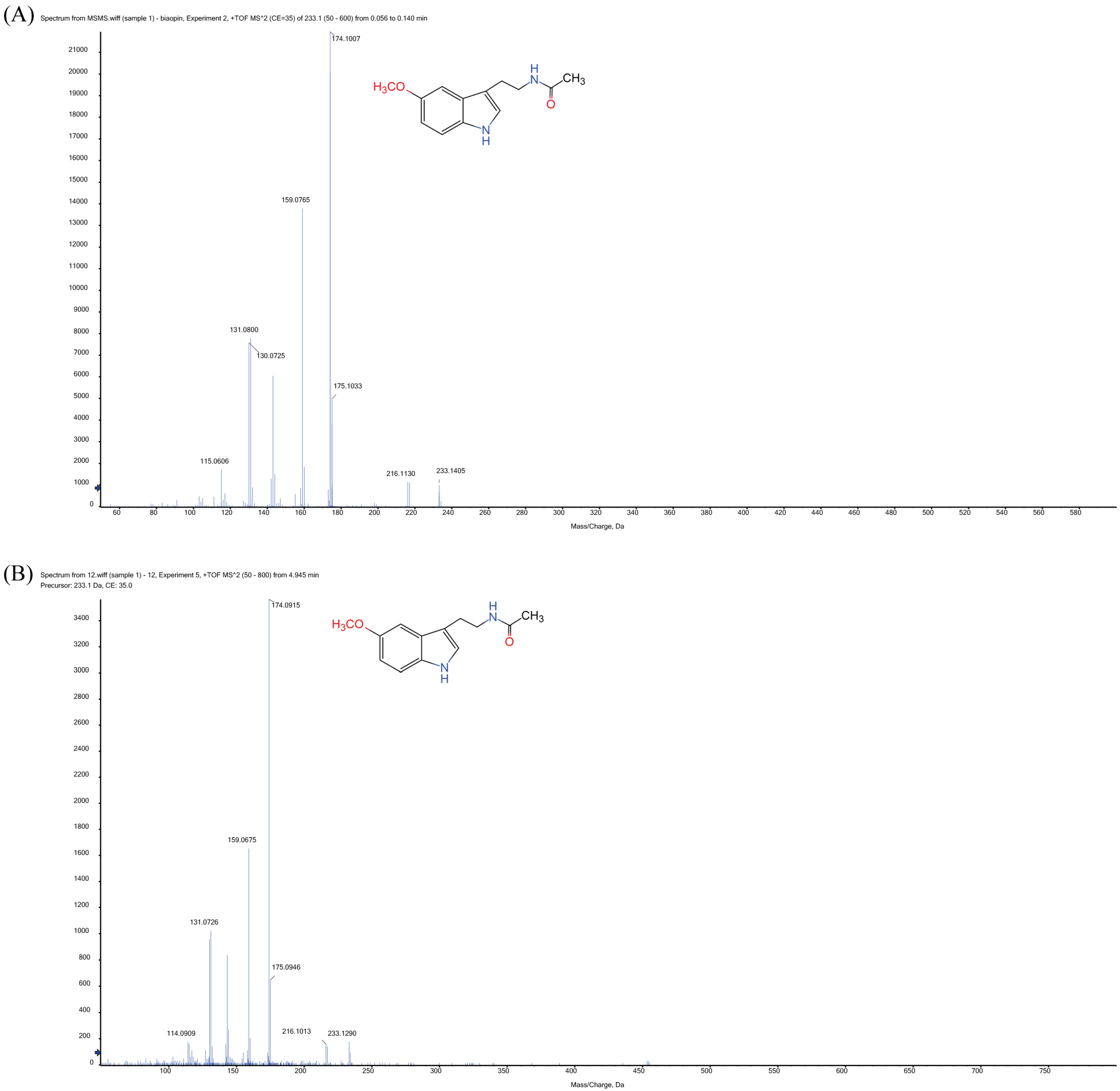
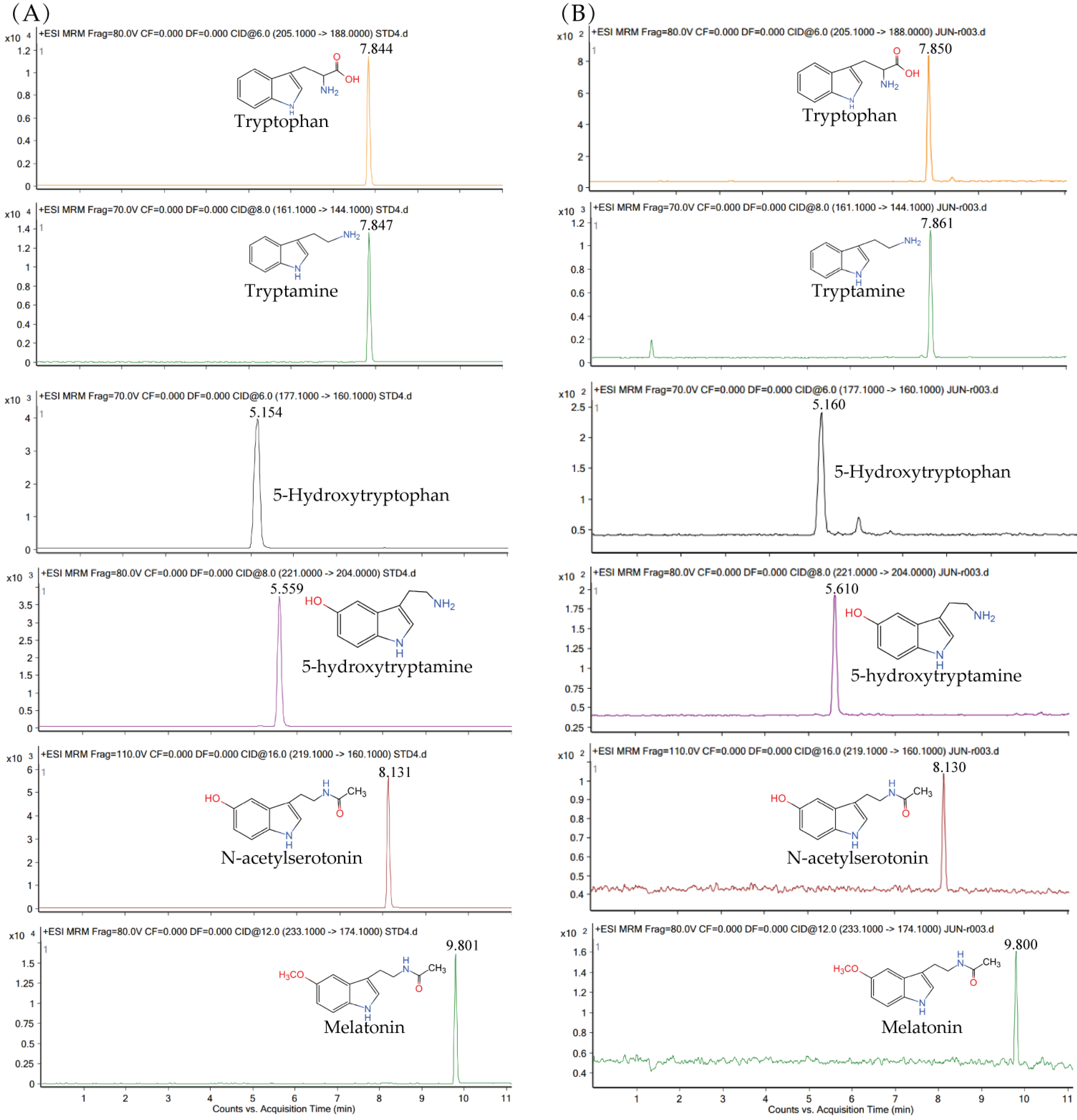
3.1. Identification of MT and MT’s Synthetic Intermediates in Xoo Extracts
3.2. Phylogenetic Analysis of xoSNAT3
3.3. Enzymatic Activity Analysis of xoSNAT3
3.4. Location of xoSNAT3 in Tobacco Cells
3.5. Role of xoSNAT3 in MT Biosynthesis in Xoo
3.6. Role of xoSNAT3 in Xoo Pathogenicity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Manchester, L.C.; Coto-Montes, A.; Boga, J.A.; Andersen, L.P.; Zhou, Z.; Galano, A.; Vriend, J.; Tan, D.X.; Reiter, R.J. Melatonin: An ancient molecule that makes oxygen metabolically tolerable. J. Pineal Res. 2015, 59, 403–419. [Google Scholar] [CrossRef] [PubMed]
- Lerner, A.B.; Case, J.D.; Takahashi, Y.; Lee, T.H.; Mori, W. Isolation of melatonin, the pineal gland factor that lightens melanocytes. J. Am. Chem. Soc. 1958, 80, 2587. [Google Scholar] [CrossRef]
- Hattori, A.; Migitaka, H.; Iigo, M.; Itoh, M.; Yamamoto, K.; Ohtani-Kaneko, R.; Hara, M.; Suzuki, T.; Reiter, R.J. Identification of melatonin in plants and its effects on plasma melatonin levels and binding to melatonin receptors in vertebrates. Biochem. Mol. Biol. Int. 1995, 35, 627–634. [Google Scholar] [PubMed]
- Manchester, L.C.; Poeggeler, B.; Alvares, F.L.; Ogden, G.B.; Reiter, R.J. Melatonin immunoreactivity in the photosynthetic prokaryote Rhodospirillum rubrum: Implications for an ancient antioxidant system. Cell. Mol. Biol. Res. 1995, 41, 391–395. [Google Scholar] [PubMed]
- Zisapel, N. New perspectives on the role of melatonin in human sleep, circadian rhythms and their regulation. Br. J. Pharmacol. 2018, 175, 3190–3199. [Google Scholar] [CrossRef] [Green Version]
- Tan, D.X.; Reiter, R.J.; Manchester, L.C.; Yan, M.T.; El-Sawi, M.; Sainz, R.M.; Mayo, J.C.; Kohen, R.; Allegra, M.; Hardeland, R. Chemical and physical properties and potential mechanisms: Melatonin as a broad spectrum antioxidant and free radical scavenger. Curr. Top. Med. Chem. 2002, 2, 181–197. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Liu, L.; Wang, L.; Li, B.; Jin, C.; Lin, X. Melatonin: A master regulator of plant development and stress responses. J. Integr. Plant Biol. 2021, 63, 126–145. [Google Scholar] [CrossRef]
- Wei, J.; Li, D.X.; Zhang, J.R.; Shan, C.; Rengel, Z.; Song, Z.B.; Chen, Q. Phytomelatonin receptor PMTR1-mediated signaling regulates stomatal closure in Arabidopsis thaliana. J. Pineal Res. 2018, 65, e12500. [Google Scholar] [CrossRef]
- Zhao, D.; Yu, Y.; Shen, Y.; Liu, Q.; Zhao, Z.; Sharma, R.; Reiter, R.J. Melatonin synthesis and function: Evolutionary history in animals and plants. Front. Endocrinol. 2019, 10, 249. [Google Scholar] [CrossRef] [Green Version]
- Byeon, Y.; Lee, H.Y.; Back, K. Cloning and characterization of the serotonin N-acetyltransferase-2 gene (SNAT2) in rice (Oryza sativa). J. Pineal Res. 2016, 61, 198–207. [Google Scholar] [CrossRef]
- Byeon, Y.; Lee, H.Y.; Back, K. Chloroplastic and cytoplasmic overexpression of sheep serotonin N-acetyltransferase in transgenic rice plants is associated with low melatonin production despite high enzyme activity. J. Pineal Res. 2015, 58, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Byeon, Y.; Lee, H.Y.; Lee, K.; Back, K. A rice chloroplast transit peptide sequence does not alter the cytoplasmic localization of sheep serotonin N-acetyltransferase expressed in transgenic rice plants. J. Pineal Res. 2014, 57, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Arnao, M.B.; Hernandez-Ruiz, J. Functions of melatonin in plants: A review. J. Pineal Res. 2015, 59, 133–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Back, K.; Tan, D.X.; Reiter, R.J. Melatonin biosynthesis in plants: Multiple pathways catalyze tryptophan to melatonin in the cytoplasm or chloroplasts. J. Pineal Res. 2016, 61, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Byeon, Y.; Lee, H.Y.; Lee, K.; Park, S.; Back, K. Cellular localization and kinetics of the rice melatonin biosynthetic enzymes SNAT and ASMT. J. Pineal Res. 2014, 56, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.; Kang, S.; Lee, K.; Park, M.; Back, K. Enzymatic features of serotonin biosynthetic enzymes and serotonin biosynthesis in plants. Plant Signal Behav. 2008, 3, 389–390. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.; Kang, K.; Lee, K.; Back, K. Characterization of rice tryptophan decarboxylases and their direct involvement in serotonin biosynthesis in transgenic rice. Planta 2007, 227, 263–272. [Google Scholar] [CrossRef]
- Tilden, A.R.; Becker, M.A.; Amma, L.L.; Arciniega, J.; McGaw, A.K. Melatonin production in an aerobic photosynthetic bacterium: An evolutionarily early association with darkness. J. Pineal Res. 1997, 22, 102–106. [Google Scholar] [CrossRef]
- Ma, Y.; Jiao, J.; Fan, X.; Sun, H.; Zhang, Y.; Jiang, J.; Liu, C. Endophytic bacterium Pseudomonas fluorescens RG11 may transform tryptophan to melatonin and promote endogenous melatonin levels in the roots of four grape cultivars. Front. Plant Sci. 2016, 7, 2068. [Google Scholar] [CrossRef] [Green Version]
- Byeon, Y.; Lee, K.; Park, Y.I.; Park, S.; Back, K. Molecular cloning and functional analysis of serotonin N-acetyltransferase from the cyanobacterium Synechocystis sp. PCC 6803. J. Pineal Res. 2013, 55, 371–376. [Google Scholar] [CrossRef]
- Chen, X.; Sun, C.; Laborda, P.; Zhao, Y.; Palmer, I.; Fu, Z.Q.; Qiu, J.; Liu, F. Melatonin treatment inhibits the growth of Xanthomonas oryzae pv. oryzae. Front. Microbiol. 2018, 9, 2280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Laborda, P.; Liu, F. Exogenous melatonin enhances rice plant resistance against Xanthomonas oryzae pv. oryzae. Plant Dis. 2020, 104, 1701–1708. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Yan, F.; Guo, W.; Zheng, H.; Lin, L.; Peng, J.; Adams, M.J.; Chen, J. Garlic virus X 11-kDa protein granules move within the cytoplasm and traffic a host protein normally found in the nucleolus. Mol. Plant Pathol. 2011, 12, 666–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kauffman, H.E.; Reddy, A.P.K.; Hsieh, S.P.Y.; Merca, S.D. An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis. Rep. 1973, 57, 537–541. [Google Scholar]
- Chen, X.; Sun, C.; Laborda, P.; He, Y.; Zhao, Y.; Li, C.; Liu, F. Melatonin treatments reduce the pathogenicity and inhibit the growth of Xanthomonas oryzae pv. oryzicola. Plant Pathol. 2019, 68, 288–296. [Google Scholar] [CrossRef]
- Pan, X.; Lu, L.; Cai, Y.D. Predicting protein subcellular location with network embedding and enrichment features. Biochim. Biophys. Acta Proteins Proteom. 2020, 1868, 140477. [Google Scholar] [CrossRef]
- Sturtz, M.; Cerezo, A.B.; Cantos-Villar, E.; Garcia-Parrilla, M.C. Determination of the melatonin content of different varieties of tomatoes (Lycopersicon esculentum) and strawberries (Fragaria ananassa). Food Chem. 2011, 127, 1329–1334. [Google Scholar] [CrossRef]
- Carter, M.D.; Calcutt, M.W.; Malow, B.A.; Rose, K.L.; Hachey, D.L. Quantitation of melatonin and n-acetylserotonin in human plasma by nanoflow LC-MS/MS and electrospray LC-MS/MS. J. Mass Spectrom. 2012, 47, 277–285. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Wang, Y.; Jin, Y.; Liu, S.; Xu, H.; Lu, X. Rapid and sensitive analysis of melatonin by LC-MS/MS and its application to pharmacokinetic study in dogs. Asian J. Pharm. 2015, 11, 273–280. [Google Scholar] [CrossRef] [Green Version]
- Ackermann, K.; Stehle, J.H. Melatonin synthesis in the human pineal gland: Advantages, implications, and difficulties. Chronobiol. Int. 2006, 23, 369–379. [Google Scholar] [CrossRef]
- Tan, D.X.; Manchester, L.C.; Qin, L.; Reiter, R.J. Melatonin: A mitochondrial targeting molecule involving mitochondrial protection and dynamics. Int. J. Mol. Sci. 2016, 17, 2124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zückert, W.R.; Marquis, H.; Goldfine, H. Modulation of enzymatic activity and biological function of Listeria monocytogenes broad-range phospholipase C by amino acid substitutions and by replacement with the Bacillus cereus ortholog. Infect. Immun. 1998, 66, 4823–4831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, L.; Zhou, Y.; Xu, Y.; Zhang, Y.; Liu, X.; Liu, B.; Chen, X.; Guo, Y.; Zeng, Z.; Zhao, Y. Structural and molecular dynamics analysis of plant serotonin N-acetyltransferase reveal an acid/base-assisted catalysis in melatonin biosynthesis. Angew. Chem. Int. Ed. 2021, 60, 12020–12026. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, J.; Gastel, J.; Klein, D.C.; Cole, P.A. Kinetic analysis of the catalytic mechanism of serotonin N-acetyltransferase (EC 2.3.1.87). J. Biol. Chem. 1998, 273, 3045–3050. [Google Scholar] [CrossRef] [Green Version]
- Kang, K.; Lee, K.; Park, S.; Byeon, Y.; Back, K. Molecular cloning of rice serotonin N-acetyltransferase, the penultimate gene in plant melatonin biosynthesis. J. Pineal Res. 2013, 55, 7–13. [Google Scholar] [CrossRef]


| No | Protein | Accession NO (NCBI) | AA Length | Functional Annotation | Specie |
|---|---|---|---|---|---|
| 1 | xoSNAT1 | WP_027703221.1 | 161 | N-acetyltransferase | Xanthomonas (Microorganism) |
| 2 | xoSNAT2 | WP_011258206.1 | 179 | N-acetyltransferase | Xanthomonas (Microorganism) |
| 3 | xoSNAT3 | WP_027703680.1 | 166 | N-acetyltransferase | Xanthomonas (Microorganism) |
| 4 | OsSNAT1 | gi|75106929 | 254 | Serotonin N-acetyltransferase | Oryza sativa (Plant) |
| 5 | OsSNAT2 | gi|75133110 | 200 | Serotonin N-acetyltransferase | Oryza sativa (Plant) |
| 6 | PhSNAT1 | XP_005526849.1 | 205 | Serotonin N-acetyltransferase | Pseudopodoces humilis (Animal) |
| 7 | MmSNAT1 | gi|11387071 | 205 | Serotonin N-acetyltransferase | Mus musculus (Animal) |
| 8 | HsSNAT1 | gi|11387096 | 207 | Serotonin N-acetyltransferase | Homo sapiens (Animal) |
| 9 | OaSNAT1 | gi|11387097 | 207 | Serotonin N-acetyltransferase | Ovis aries (Animal) |
| 10 | SaSNAT1 | gi|827410280 | 142 | N-acetyltransferase | Streptococcus agalactiae (Microorganism) |
| NO | Standard MT | MT from Xoo Extracts |
|---|---|---|
| 1 | 174.1007 | 174.0915 |
| 2 | 159.0765 | 159.0675 |
| 3 | 131.0800 | 131.0726 |
| 4 | 175.1033 | 175.0946 |
| 5 | 115.0606 | 114.0909 |
| 6 | 233.1405 | 233.1290 |
| 7 | 216.1130 | 216.1013 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Zhao, Y.; Laborda, P.; Yang, Y.; Liu, F. Molecular Cloning and Characterization of a Serotonin N-Acetyltransferase Gene, xoSNAT3, from Xanthomonas oryzae pv. oryzae. Int. J. Environ. Res. Public Health 2023, 20, 1865. https://doi.org/10.3390/ijerph20031865
Chen X, Zhao Y, Laborda P, Yang Y, Liu F. Molecular Cloning and Characterization of a Serotonin N-Acetyltransferase Gene, xoSNAT3, from Xanthomonas oryzae pv. oryzae. International Journal of Environmental Research and Public Health. 2023; 20(3):1865. https://doi.org/10.3390/ijerph20031865
Chicago/Turabian StyleChen, Xian, Yancun Zhao, Pedro Laborda, Yong Yang, and Fengquan Liu. 2023. "Molecular Cloning and Characterization of a Serotonin N-Acetyltransferase Gene, xoSNAT3, from Xanthomonas oryzae pv. oryzae" International Journal of Environmental Research and Public Health 20, no. 3: 1865. https://doi.org/10.3390/ijerph20031865







