Bimodal Imaging of Tumors via Genetically Engineered Escherichia coli
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Plasmid Construction
2.2. Transformation and Construction of GVs-miRFP680 MG1655
2.3. Bacterial Culture and Expression of GVs-miRFP680 MG1655
2.4. Characteristics of Gas Vesicles in GVs-miRFP680 MG1655
2.5. Cell Culture
2.6. Animal Models
2.7. Ultrasound Imaging Capability of GVs-miRFP680 MG1655
2.8. Optical Imaging Capability of GVs-miRFP680 MG1655
2.9. Statistical Analysis
3. Results
3.1. Construction of GVs-miRFP680 MG1655 Bacteria
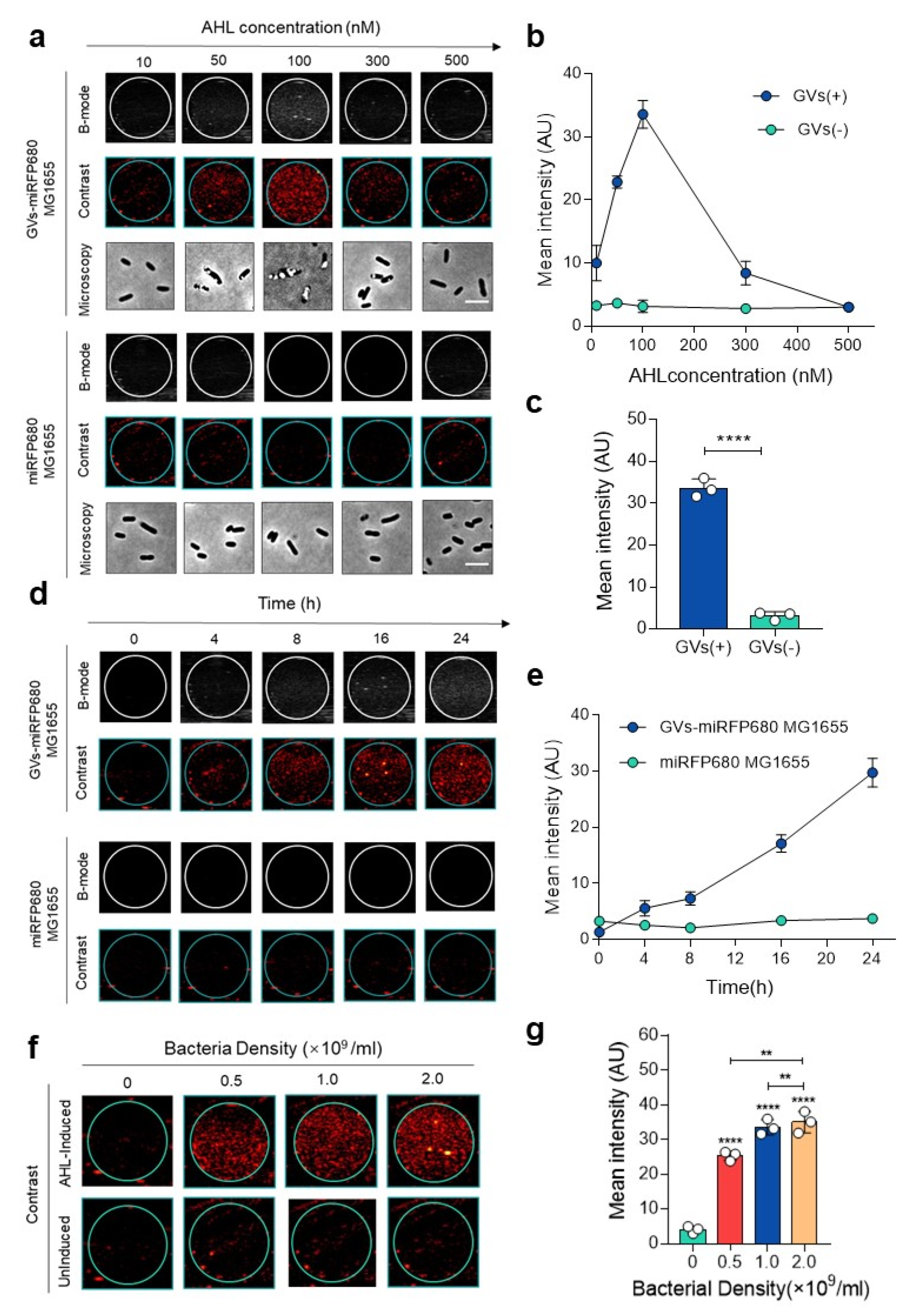
3.2. In Vitro Ultrasound Imaging of GVs-miRFP680 MG1655
3.3. In Vivo Ultrasound Imaging of GVs-miRFP680 MG1655
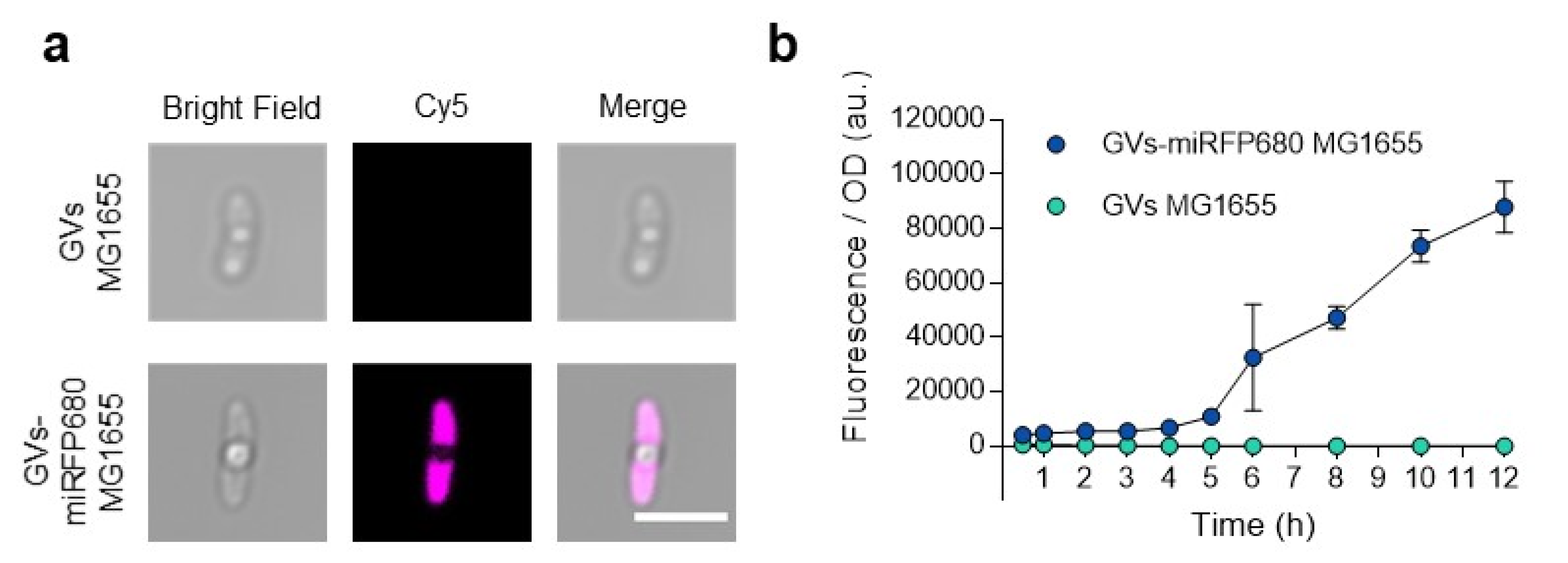
3.4. In Vitro Optical Imaging of GVs-miRFP680 MG1655
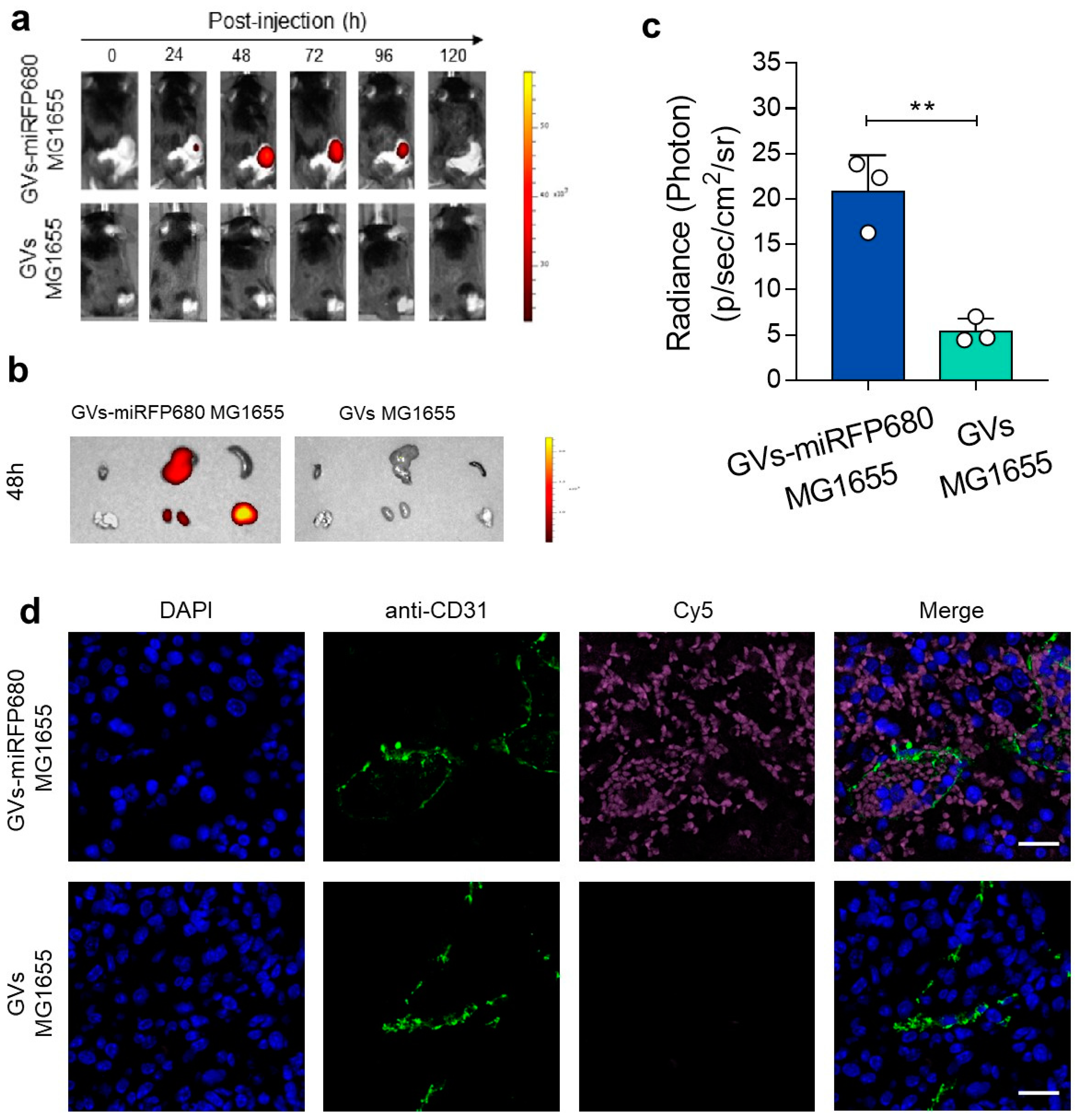
3.5. In Vivo Tracking Capability of the Tumor-Homing Characteristic of GVs-miRFP680 MG1655
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dougan, M.; Dougan, S.K. Programmable bacteria as cancer therapy. Nat. Med. 2019, 25, 1030–1031. [Google Scholar] [CrossRef] [PubMed]
- Forbes, N.S. Engineering the perfect (bacterial) cancer therapy. Nat. Rev. Cancer 2010, 10, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Gravekamp, C.; Bermudes, D.; Liu, K. Tumour-targeting bacteria engineered to fight cancer. Nat. Rev. Cancer 2018, 18, 727–743. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Castro, S.; Coker, C.; Hinchliffe, T.E.; Arpaia, N.; Danino, T. Programmable bacteria induce durable tumor regression and systemic antitumor immunity. Nat. Med. 2019, 25, 1057–1063. [Google Scholar]
- Zhang, Y.; Ji, W.; He, L.; Chen, Y.; Ding, X.; Sun, Y.; Hu, S.; Yang, H.; Huang, W.; Zhang, Y.; et al. E. coli Nissle 1917-Derived Minicells for Targeted Delivery of Chemotherapeutic Drug to Hypoxic Regions for Cancer Therapy. Theranostics 2018, 8, 1690–1705. [Google Scholar]
- Park, S.H.; Zheng, J.H.; Nguyen, V.H.; Jiang, S.N.; Kim, D.Y.; Szardenings, M.; Min, J.H.; Hong, Y.; Choy, H.E.; Min, J.J. RGD Peptide Cell-Surface Display Enhances the Targeting and Therapeutic Efficacy of Attenuated Salmonella-mediated Cancer Therapy. Theranostics 2016, 6, 1672–1682. [Google Scholar] [CrossRef]
- Gao, R.; Liu, F.; Liu, W.; Zeng, S.; Chen, J.; Gao, R.; Wang, L.; Fang, C.; Song, L.; Sedgwick, A.C.; et al. Background-suppressed tumor-targeted photoacoustic imaging using bacterial carriers. Proc. Natl. Acad. Sci. USA 2022, 119, e2121982119. [Google Scholar] [CrossRef]
- Min, J.J.; Nguyen, V.H.; Kim, H.J.; Hong, Y.; Choy, H.E. Quantitative bioluminescence imaging of tumor-targeting bacteria in living animals. Nat. Protoc. 2008, 3, 629–636. [Google Scholar] [CrossRef]
- Leschner, S.; Westphal, K.; Dietrich, N.; Viegas, N.; Jablonska, J.; Lyszkiewicz, M.; Lienenklaus, S.; Falk, W.; Gekara, N.; Loessner, H.; et al. Tumor invasion of Salmonella enterica serovar Typhimurium is accompanied by strong hemorrhage promoted by TNF-alpha. PLoS ONE 2009, 4, e6692. [Google Scholar] [CrossRef]
- Jiang, S.N.; Phan, T.X.; Nam, T.K.; Nguyen, V.H.; Kim, H.S.; Bom, H.S.; Choy, H.E.; Hong, Y.; Min, J.J. Inhibition of tumor growth and metastasis by a combination of Escherichia coli-mediated cytolytic therapy and radiotherapy. Mol. Ther. 2010, 18, 635–642. [Google Scholar] [CrossRef]
- Wang, C.; Wang, Z.; Zhao, T.; Li, Y.; Huang, G.; Sumer, B.D.; Gao, J. Optical molecular imaging for tumor detection and image-guided surgery. Biomaterials 2018, 157, 62–75. [Google Scholar] [CrossRef] [PubMed]
- Bourdeau, R.W.; Lee-Gosselin, A.; Lakshmanan, A.; Farhadi, A.; Kumar, S.R.; Nety, S.P.; Shapiro, M.G. Acoustic reporter genes for noninvasive imaging of microorganisms in mammalian hosts. Nature 2018, 553, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Hurt, R.C.; Buss, M.T.; Wong, K.; Sawyer, D.P.; Swift, M.B.; Dutka, P.; Mittelstein, D.R.; Jin, Z.; Abedi, M.H.; Deshpande, R.; et al. Genomically Mined Acoustic Reporter Genes Enable On-Demand In Vivo Monitoring of Tumor-Homing Bacteria. bioRxiv 2021. [Google Scholar] [CrossRef]
- Bar-Zion, A.; Nourmahnad, A.; Mittelstein, D.R.; Shivaei, S.; Yoo, S.; Buss, M.T.; Hurt, R.C.; Malounda, D.; Abedi, M.H.; Lee-Gosselin, A.; et al. Acoustically triggered mechanotherapy using genetically encoded gas vesicles. Nat. Nanotechnol. 2021, 16, 1403–1412. [Google Scholar] [CrossRef] [PubMed]
- Matlashov, M.E.; Shcherbakova, D.M.; Alvelid, J.; Baloban, M.; Pennacchietti, F.; Shemetov, A.A.; Testa, I.; Verkhusha, V.V. A set of monomeric near-infrared fluorescent proteins for multicolor imaging across scales. Nat. Commun. 2020, 11, 239. [Google Scholar] [CrossRef]
- Weissleder, R.; Ntziachristos, V. Shedding light onto live molecular targets. Nat. Med. 2003, 9, 123–128. [Google Scholar] [CrossRef]
- Shcherbakova, D.M.; Baloban, M.; Verkhusha, V.V. Near-infrared fluorescent proteins engineered from bacterial phytochromes. Curr. Opin. Chem. Biol. 2015, 27, 52–63. [Google Scholar] [CrossRef]
- Shcherbakova, D.M.; Baloban, M.; Emelyanov, A.V.; Brenowitz, M.; Guo, P.; Verkhusha, V.V. Bright monomeric near-infrared fluorescent proteins as tags and biosensors for multiscale imaging. Nat. Commun. 2016, 7, 12405. [Google Scholar] [CrossRef]
- Tiwari, D.K.; Tiwari, M.; Jin, T. Near-infrared fluorescent protein and bioluminescence-based probes for high-resolution in vivo optical imaging. Mater. Adv. 2020, 1, 967–987. [Google Scholar]
- Kang, S.R.; Jo, E.J.; Nguyen, V.H.; Zhang, Y.; Yoon, H.S.; Pyo, A.; Kim, D.Y.; Hong, Y.; Bom, H.S.; Min, J.J. Imaging of tumor colonization by Escherichia coli using (18)F-FDS PET. Theranostics 2020, 10, 4958–4966. [Google Scholar] [CrossRef]
- Abedi, M.H.; Yao, M.S.; Mittelstein, D.R.; Bar-Zion, A.; Swift, M.B.; Lee-Gosselin, A.; Barturen-Larrea, P.; Buss, M.T.; Shapiro, M.G. Ultrasound-controllable engineered bacteria for cancer immunotherapy. Nat. Commun. 2022, 13, 1585. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Jiang, F.; Ji, X.; Wang, L.; Wang, Y.; Zhang, L.; Tang, Y.; Wang, D.; Luo, Y.; Li, N.; et al. Genetically Engineered Bacterial Protein Nanoparticles for Targeted Cancer Therapy. Int. J. Nanomed. 2021, 16, 105–117. [Google Scholar] [CrossRef]
- Pinero-Lambea, C.; Bodelon, G.; Fernandez-Perianez, R.; Cuesta, A.M.; Alvarez-Vallina, L.; Fernandez, L.A. Programming controlled adhesion of E. coli to target surfaces, cells, and tumors with synthetic adhesins. ACS Synth. Biol. 2015, 4, 463–473. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Wang, Y.; Li, D.; Wang, L.; Li, Z.; Yan, F. Bimodal Imaging of Tumors via Genetically Engineered Escherichia coli. Pharmaceutics 2022, 14, 1804. https://doi.org/10.3390/pharmaceutics14091804
Zhang L, Wang Y, Li D, Wang L, Li Z, Yan F. Bimodal Imaging of Tumors via Genetically Engineered Escherichia coli. Pharmaceutics. 2022; 14(9):1804. https://doi.org/10.3390/pharmaceutics14091804
Chicago/Turabian StyleZhang, Linlin, Yuanyuan Wang, Dengjin Li, Liang Wang, Zhenzhou Li, and Fei Yan. 2022. "Bimodal Imaging of Tumors via Genetically Engineered Escherichia coli" Pharmaceutics 14, no. 9: 1804. https://doi.org/10.3390/pharmaceutics14091804





