Forest Above-Ground Biomass Inversion Using Optical and SAR Images Based on a Multi-Step Feature Optimized Inversion Model
Abstract
:1. Introduction
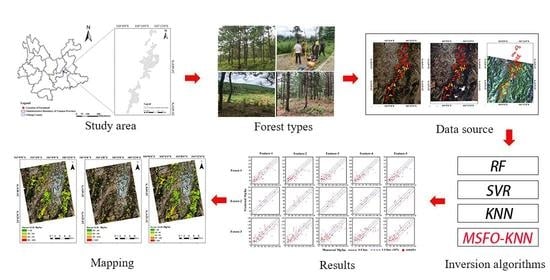
2. Materials
2.1. Test Site
2.2. Satellite Data and Preprocessing
2.2.1. Optical Data Acquisitions and Preprocessing
2.2.2. Feature Extraction from Optical and SAR Images
2.3. Ground-Measured Forest AGB
3. Methodology
3.1. Random Forest (RF) for Original Remote Sensing Feature Optimization
3.2. Feature Combination Obtained with a Fast Iterative Procedure Embedded in KNN Algorithms
- (1)
- The trained datasets comprised of the field samples and RF selected RS variables are defined as , where describes the number of selected original RS variables after the ordering using the RF variable importance function. Among them, , is the pixel value of the feature character at the plot, where is the plot number and means the transform of the vector.
- (2)
- Initializing the original best optimized feature subset as , and the root means square error (RMSE) for the best estimation model as , it is initialized according to the maximum AGB value at the study area.
- (3)
- Generate KNN models and their retrieval RMSE values by feature combinations of , where . The RMSE values are calculated with the leave one out cross validation (LOOCV) method.
- (4)
- The lowest value of RMSE calculated in step 3) is set as ; if , then and the feature combination for the is set for .
- (5)
- Step 3 is repeated iterative times until .
3.3. Forest Biomass Estimation and Validation Using Proposed MSFO-KNN
4. Results
4.1. Remote Sensing Features Optimization Analysis
4.1.1. Original Feature Ordering and Optimization by RF
4.1.2. Second Step Feature Optimization by the Fast Iterative Procedure
4.1.3. Forest AGB Retrievals Using Different Nonparametric Algorithms
5. Discussion
5.1. Capabilities and Limitations of the Features Extracted from Optical and SAR Images
5.2. Differences between Forest Types on AGB Estimation
5.3. Performance of the Proposed MSFO-KNN Algorithm
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stocker, T.; Qin, D.; Plattner, G.-K.; Tignor, M.; Allen, S.K.; Boschung, J.; Nauels, A.; Xia, Y.; Bex, V.; Midgley, P.M.; et al. Climate Change 2013—The Physical Science Basis: Working Group I Contribution to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; IPCC, Ed.; Cambridge University Press: Cambridge, UK, 2014; ISBN 978-1-107-41532-4. [Google Scholar]
- Le Toan, T.; Quegan, S.; Davidson, M.W.J.; Balzter, H.; Paillou, P.; Papathanassiou, K.; Plummer, S.; Rocca, F.; Saatchi, S.; Shugart, H.; et al. The BIOMASS mission: Mapping global forest biomass to better understand the terrestrial carbon cycle. Remote Sens. Environ. 2011, 115, 2850–2860. [Google Scholar] [CrossRef] [Green Version]
- Soja, M.J.; Quegan, S.; d’Alessandro, M.M.; Banda, F.; Scipal, K.; Tebaldini, S.; Ulander, L.M.H. Mapping above-ground biomass in tropical forests with ground-cancelled P-band SAR and limited reference data. Remote Sens. Environ. 2021, 253, 112153. [Google Scholar] [CrossRef]
- Hayashi, M.; Motohka, T.; Sawada, Y. Aboveground Biomass Mapping Using ALOS-2/PALSAR-2 Time-Series Images for Borneo’s Forest. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2019, 12, 5167–5177. [Google Scholar] [CrossRef]
- Lu, D.; Chen, Q.; Wang, G.; Moran, E.; Batistella, M.; Zhang, M.; Vaglio Laurin, G.; Saah, D. Aboveground Forest Biomass Estimation with Landsat and LiDAR Data and Uncertainty Analysis of the Estimates. Int. J. For. Res. 2012, 2012, 436537. [Google Scholar] [CrossRef]
- Gao, Y.; Lu, D.; Li, G.; Wang, G.; Chen, Q.; Liu, L.; Li, D. Comparative Analysis of Modeling Algorithms for Forest Aboveground Biomass Estimation in a Subtropical Region. Remote Sens. 2018, 10, 627. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Veiga, P.; Quegan, S.; Carreiras, J.; Persson, H.J.; Fransson, J.E.S.; Hoscilo, A.; Ziółkowski, D.; Stereńczak, K.; Lohberger, S.; Stängel, M.; et al. Forest biomass retrieval approaches from earth observation in different biomes. Int. J. Appl. Earth Obs. Geoinf. 2019, 77, 53–68. [Google Scholar] [CrossRef]
- Chowdhury, T.; Thiel, C.; Schmullius, C.; Stelmaszczuk-Górska, M. Polarimetric Parameters for Growing Stock Volume Estimation Using ALOS PALSAR L-Band Data over Siberian Forests. Remote Sens. 2013, 5, 5725–5756. [Google Scholar] [CrossRef] [Green Version]
- Foody, G.M.; Boyd, D.S.; Cutler, M.E.J. Predictive relations of tropical forest biomass from Landsat TM data and their transferability between regions. Remote Sens. Environ. 2003, 85, 463–474. [Google Scholar] [CrossRef]
- Mitchard, E.T.A.; Saatchi, S.S.; Lewis, S.L.; Feldpausch, T.R.; Woodhouse, I.H.; Sonké, B.; Rowland, C.; Meir, P. Measuring biomass changes due to woody encroachment and deforestation/degradation in a forest–savanna boundary region of central Africa using multi-temporal L-band radar backscatter. Remote Sens. Environ. 2011, 115, 2861–2873. [Google Scholar] [CrossRef] [Green Version]
- Godinho Cassol, H.L.; De Oliveira E Cruz De Aragão, L.E.; Moraes, E.C.; De Brito Carreiras, J.M.; Shimabukuro, Y.E. Quad-pol advanced land observing satellite/phased array L-band synthetic aperture radar-2 (ALOS/PALSAR-2) data for modelling secondary forest above-ground biomass in the central Brazilian Amazon. Int. J. Remote Sens. 2021, 42, 4985–5009. [Google Scholar] [CrossRef]
- Montesano, P.M.; Cook, B.D.; Sun, G.; Simard, M.; Nelson, R.F.; Ranson, K.J.; Zhang, Z.; Luthcke, S. Achieving accuracy requirements for forest biomass mapping: A spaceborne data fusion method for estimating forest biomass and LiDAR sampling error. Remote Sens. Environ. 2013, 130, 153–170. [Google Scholar] [CrossRef]
- Li, Y.; Li, M.; Li, C.; Liu, Z. Forest aboveground biomass estimation using Landsat 8 and Sentinel-1A data with machine learning algorithms. Sci. Rep. 2020, 10, 9952. [Google Scholar] [CrossRef] [PubMed]
- Lu, D. The potential and challenge of remote sensing-based biomass estimation. Int. J. Remote Sens. 2006, 27, 1297–1328. [Google Scholar] [CrossRef]
- Ou, G.; Li, C.; Lv, Y.; Wei, A.; Xiong, H.; Xu, H.; Wang, G. Improving Aboveground Biomass Estimation of Pinus densata Forests in Yunnan Using Landsat 8 Imagery by Incorporating Age Dummy Variable and Method Comparison. Remote Sens. 2019, 11, 738. [Google Scholar] [CrossRef] [Green Version]
- Lu, D.; Chen, Q.; Wang, G.; Liu, L.; Li, G.; Moran, E. A survey of remote sensing-based aboveground biomass estimation methods in forest ecosystems. Int. J. Digit. Earth 2016, 9, 63–105. [Google Scholar] [CrossRef]
- Sandberg, G.; Ulander, L.M.H.; Fransson, J.E.S.; Holmgren, J.; Le Toan, T. L- and P-band backscatter intensity for biomass retrieval in hemiboreal forest. Remote Sens. Environ. 2011, 115, 2874–2886. [Google Scholar] [CrossRef]
- Chen, L.; Wang, Y.; Ren, C.; Zhang, B.; Wang, Z. Assessment of multi-wavelength SAR and multispectral instrument data for forest aboveground biomass mapping using random forest kriging. For. Ecol. Manag. 2019, 447, 12–25. [Google Scholar] [CrossRef]
- Ji, Y.; Xu, K.; Zeng, P.; Zhang, W. GA-SVR Algorithm for Improving Forest Above Ground Biomass Estimation Using SAR Data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2021, 14, 6585–6595. [Google Scholar] [CrossRef]
- Feng, Y.; Lu, D.; Chen, Q.; Keller, M.; Moran, E.; dos-Santos, M.N.; Bolfe, E.L.; Batistella, M. Examining effective use of data sources and modeling algorithms for improving biomass estimation in a moist tropical forest of the Brazilian Amazon. Int. J. Digit. Earth 2017, 10, 996–1016. [Google Scholar] [CrossRef]
- Yang, Z.; Shao, Y.; Li, K.; Liu, Q.; Liu, L.; Brisco, B. An improved scheme for rice phenology estimation based on time-series multispectral HJ-1A/B and polarimetric RADARSAT-2 data. Remote Sens. Environ. 2017, 195, 184–201. [Google Scholar] [CrossRef]
- Fleming, A.L.; Wang, G.; McRoberts, R.E. Comparison of methods toward multi-scale forest carbon mapping and spatial uncertainty analysis: Combining national forest inventory plot data and landsat TM images. Eur. J. For. Res. 2015, 134, 125–137. [Google Scholar] [CrossRef]
- Breidenbach, J.; Næsset, E.; Gobakken, T. Improving k-nearest neighbor predictions in forest inventories by combining high and low density airborne laser scanning data. Remote Sens. Environ. 2012, 117, 358–365. [Google Scholar] [CrossRef]
- McRoberts, R.E.; Chen, Q.; Domke, G.M.; Næsset, E.; Gobakken, T.; Chirici, G.; Mura, M. Optimizing nearest neighbour configurations for airborne laser scanning-assisted estimation of forest volume and biomass. Forestry 2017, 90, 99–111. [Google Scholar] [CrossRef]
- Tian, X.; Su, Z.; Chen, E.; Li, Z.; van der Tol, C.; Guo, J.; He, Q. Estimation of forest above-ground biomass using multi-parameter remote sensing data over a cold and arid area. Int. J. Appl. Earth Obs. Geoinf. 2012, 14, 160–168. [Google Scholar] [CrossRef]
- Han, Z.; Jiang, H.; Wang, W.; Li, Z.; Chen, E.; Yan, M.; Tian, X. Forest Above-Ground Biomass Estimation Using KNN-FIFS Method Based on Multi-Source Remote Sensing Data. Sci. Silvae Sin. 2018, 54, 70–79. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, W.; Cui, Y. Retrieval of Forest Aboveground biomass from optical and SAR data supported by parameter optimization. J. Beijing For. Univ. 2020, 42, 11–19. [Google Scholar]
- Bu, F. Estimation of Forest Aboveground Biomass Based on Multi-Source Remote Sensing Data; Nanjing University of Information Engineering: Nanjing, China, 2019. [Google Scholar]
- Jensen, J.R.; Lulla, K. Introductory digital image processing: A remote sensing perspective. Geocarto Int. 1987, 2, 65. [Google Scholar] [CrossRef]
- Freeman, A.; Durden, S.L. A three-component scattering model for polarimetric SAR data. IEEE Trans. Geosci. Remote Sens. 1998, 36, 963–973. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, Y.; Moriyama, T.; Ishido, M.; Yamada, H. Four-component scattering model for polarimetric SAR image decomposition. Tech. Rep. Ieice Sane 2005, 104, 1699–1706. [Google Scholar] [CrossRef]
- Cloude, S.R.; Pottier, E. A review of target decomposition theorems in radar polarimetry. IEEE Transac. Geosci. Remote Sens. 1996, 34, 498–518. [Google Scholar] [CrossRef]
- Meng, X. Forest Mensuration; China Forestry Publishing House: Beijing, China, 2006; Volume 241–245, ISBN 978-1-4020-5990-2. [Google Scholar]
- Huang, C.; Zhang, J.; Yang, W.; Tang, X.; Zhao, A. Dynamics on forest carbon stock in Sichuan Province and Chongqing City. Acta Ecol. Sin. 2008, 28, 0966–0975. [Google Scholar]
- Belgiu, M.; Drăguţ, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Wei, J.; Fan, Y.; Yu, Y. Estimation of canopy biomass by polarimetric decomposition of GF-3 full polarimetric SAR data Forestry Science. Sci. Silvae Sin. 2020, 56, 10. [Google Scholar] [CrossRef]
- Woodhouse, I.H.; Mitchard, E.T.A.; Brolly, M.; Maniatis, D.; Ryan, C.M. Radar backscatter is not a “direct measure” of forest biomass. Nat. Clim. Chang. 2012, 2, 556–557. [Google Scholar] [CrossRef]
- Englhart, S.; Keuck, V.; Siegert, F. Modeling Aboveground Biomass in Tropical Forests Using Multi-Frequency SAR Data—A Comparison of Methods. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2012, 5, 298–306. [Google Scholar] [CrossRef]








| Parameters | Values |
|---|---|
| Polarization | HHHV, VH, VV |
| Incidence angle | 39.104° |
| Wavelength | 0.0555 m |
| Path | Ascending |
| Slant range pixel spacing | 5.12 m |
| Azimuth pixel spacing | 2.248 m |
| Vegetation Indices | Equation |
|---|---|
| Normalized Difference Vegetation Index (NDVI) | |
| Simple Ratio (SR) | |
| Visible Atmospherically Resistant Index (VARI) | |
| Differential Vegetation Index (DVI) | |
| Perpendicular Vegetation Index (PVI) | |
| Ratio Vegetation Index (RVI) | |
| Soil and Atmospherically Resistant Vegetation Index (SARVI) | |
| Enhanced Vegetation Index (EVI) | |
| Soil and Atmospherically Resistant Vegetation Index (MSARVI) | |
| Normalized Burn Ratio (NBR) | |
| Mid Infrared Index (MidIR) | |
| Moisture Stress Index (MSI) |
| Texture | Function | Interpretation |
|---|---|---|
| Mean (Me) | The local mean value of the processing window | |
| Variance (Var) | The local variance of the processing window | |
| Homogeneity (Homo) | An inverse of contrast | |
| Contrast (Con) | The amount of local variations present in an image | |
| Dissimilarity (Dis) | The absolute values of the grayscale difference | |
| Entropy (En) | A measure of the complexity of the texture | |
| Angular Second moment (Asm) | A measure of homogeneity of the image | |
| Correlation (Cor) | A measure of gray-tone linear-dependencies in the image. |
| Decomposition Method | Features |
|---|---|
| H-A-alpha | Entropy (H), anisotropy (A), Alpha angle (alpha), Eigenvalue 1 (L1), Eigenvalue 2 (L2), Beta angle (), Delta angle (), Gamma angle (), radar vegetation index (), entropy of each eigenvalue (), total backscattering power (Span). |
| Freeman 3 | Volume scattering component (Fre_Vol), double bounce scattering component (Fre_Dbl), surface scattering component (Fre_Odd), the ratio of surface scattering and volume scattering (). |
| Yamaguchi 4 | Volume scattering component (Yam_Vol), double bounce scattering component (Yam_Dbl), surface scattering component (Yam_Odd), helix scattering component (Yam_helix), the ratio of surface scat-tering and volume scattering (). |
| Forest Type | Number | AGB (Mg/ha) Above-Ground Biomass | Mean (Mg/ha) | Standard (Mg/ha) |
|---|---|---|---|---|
| Forest-1 | 55 | 9.34~135.71 | 44.90 | 28.11 |
| Forest-2 | 23 | 42.30~150.71 | 80.60 | 30.72 |
| Forest-3 | 78 | 9.34~150.34 | 55.42 | 33.17 |
| Forest Types | Feature Groups | Numbers of Chosen Feature | Numbers of Feature | Selected Features |
|---|---|---|---|---|
| Forest-1 | Feature-1 | 10 | 40 | Green, Meg, Disr, Varb, Conr |
| Feature-2 | 10 | 20 | Blue, Green, Swir2, Swir1, Red, EVI, MidIR, Corg, Ens1, Meg | |
| Feature-3 | 8 | 40 | L3, T22, T33, T23real, VVdB, H, , Mespan | |
| Feature-4 | 7 | 20 | EVI, MidIR, Red, MSI, Meg, Mes2, Cons1 | |
| Feature-5 | 5 | 30 | BlueGF1, MidIRLad, Meg_GF1, NDVILad, RVILad | |
| Forest-2 | Feature-1 | 5 | 40 | RVI, Varg, Varb, Disr, Conb |
| Feature-2 | 4 | 20 | Homos2, Corr, Ens2, Vars1 | |
| Feature-3 | 5 | 20 | T23imag, , L3, Asmspan, Dissspan, Yam_OdddB | |
| Feature-4 | 4 | 20 | Mer, Homos2, Vars2, Yam_DbldB | |
| Feature-5 | 4 | 10 | GreenGF1, Asms2_GF1, Corr_GF1, H1 | |
| Forest-3 | Feature-1 | 4 | 40 | Blue, red, Conr, Cong |
| Feature-2 | 5 | 20 | RVI, MidIR, SR, NDVI, Cons1 | |
| Feature-3 | 5 | 40 | Yam_VoldB, A, T13imag, T33dB, Conspan | |
| Feature-4 | 5 | 40 | VARI, Swir1, Ens1, Vars2, H2 | |
| Feature-5 | 3 | 10 | GreenGF1, RedGF1, RedLad |
| Forest Type | Feature Group | R2 | RMSE |
|---|---|---|---|
| Forest-1 | Feature-1 | 0.68 | 16.27 |
| Feature-2 | 0.63 | 18.05 | |
| Feature-3 | 0.57 | 18.87 | |
| Feature-4 | 0.63 | 18.25 | |
| Feature-5 | 0.70 | 16.18 | |
| Forest-2 | Feature-1 | 0.71 | 17.01 |
| Feature-2 | 0.56 | 20.96 | |
| Feature-3 | 0.70 | 18.60 | |
| Feature-4 | 0.70 | 18.27 | |
| Feature-5 | 0.72 | 17.66 | |
| Forest-3 | Feature-1 | 0.68 | 19.75 |
| Feature-2 | 0.60 | 21.79 | |
| Feature-3 | 0.60 | 21.87 | |
| Feature-4 | 0.62 | 21.34 | |
| Feature-5 | 0.71 | 18.67 |
| Method | Forest Type | Feature Group | R2 | RMSE |
|---|---|---|---|---|
| RF | Forest-1 | Feature-1 | 0.62 | 17.42 |
| Feature-2 | 0.39 | 22.01 | ||
| Feature-3 | 0.15 | 26.03 | ||
| Feature-4 | 0.33 | 23.51 | ||
| Feature-5 | 0.61 | 17.91 | ||
| Forest-2 | Feature-1 | 0.37 | 26.91 | |
| Feature-2 | 0.30 | 26.29 | ||
| Feature-3 | 0.24 | 31.65 | ||
| Feature-4 | 0.13 | 32.55 | ||
| Feature-5 | 0.27 | 28.55 | ||
| Forest-3 | Feature-1 | 0.57 | 22.09 | |
| Feature-2 | 0.33 | 28.38 | ||
| Feature-3 | 0.31 | 28.21 | ||
| Feature-4 | 0.36 | 27.37 | ||
| Feature-5 | 0.61 | 20.78 | ||
| SVR | Forest-1 | Feature-1 | 0.59 | 21.47 |
| Feature-2 | 0.39 | 26.11 | ||
| Feature-3 | 0.30 | 27.93 | ||
| Feature-4 | 0.41 | 25.83 | ||
| Feature-5 | 0.61 | 20.83 | ||
| Forest-2 | Feature-1 | 0.59 | 18.04 | |
| Feature-2 | 0.50 | 20.01 | ||
| Feature-3 | 0.36 | 22.41 | ||
| Feature-4 | 0.42 | 21.52 | ||
| Feature-5 | 0.64 | 16.91 | ||
| Forest-3 | Feature-1 | 0.27 | 26.41 | |
| Feature-2 | 0.25 | 26.79 | ||
| Feature-3 | 0.20 | 31.89 | ||
| Feature-4 | 0.32 | 25.49 | ||
| Feature-5 | 0.49 | 22.36 | ||
| KNN | Forest-1 | Feature-1 | 0.22 | 30.78 |
| Feature-2 | 0.10 | 36.60 | ||
| Feature-3 | 0.27 | 28.03 | ||
| Feature-4 | 0.08 | 33.32 | ||
| Feature-5 | 0.31 | 26.96 | ||
| Forest-2 | Feature-1 | 0.35 | 25.59 | |
| Feature-2 | 0.33 | 25.77 | ||
| Feature-3 | 0.13 | 25.79 | ||
| Feature-4 | 0.18 | 26.42 | ||
| Feature-5 | 0.48 | 22.15 | ||
| Forest-3 | Feature-1 | 0.45 | 25.70 | |
| Feature-2 | 0.29 | 29.09 | ||
| Feature-3 | 0.25 | 30.67 | ||
| Feature-4 | 0.34 | 28.50 | ||
| Feature-5 | 0.64 | 21.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Zhao, L.; Li, Y.; Shi, J.; Yan, M.; Ji, Y. Forest Above-Ground Biomass Inversion Using Optical and SAR Images Based on a Multi-Step Feature Optimized Inversion Model. Remote Sens. 2022, 14, 1608. https://doi.org/10.3390/rs14071608
Zhang W, Zhao L, Li Y, Shi J, Yan M, Ji Y. Forest Above-Ground Biomass Inversion Using Optical and SAR Images Based on a Multi-Step Feature Optimized Inversion Model. Remote Sensing. 2022; 14(7):1608. https://doi.org/10.3390/rs14071608
Chicago/Turabian StyleZhang, Wangfei, Lixian Zhao, Yun Li, Jianmin Shi, Min Yan, and Yongjie Ji. 2022. "Forest Above-Ground Biomass Inversion Using Optical and SAR Images Based on a Multi-Step Feature Optimized Inversion Model" Remote Sensing 14, no. 7: 1608. https://doi.org/10.3390/rs14071608