Recent Advances in Exosome-Based Drug Delivery for Cancer Therapy
Abstract
:Simple Summary
Abstract
1. Introduction
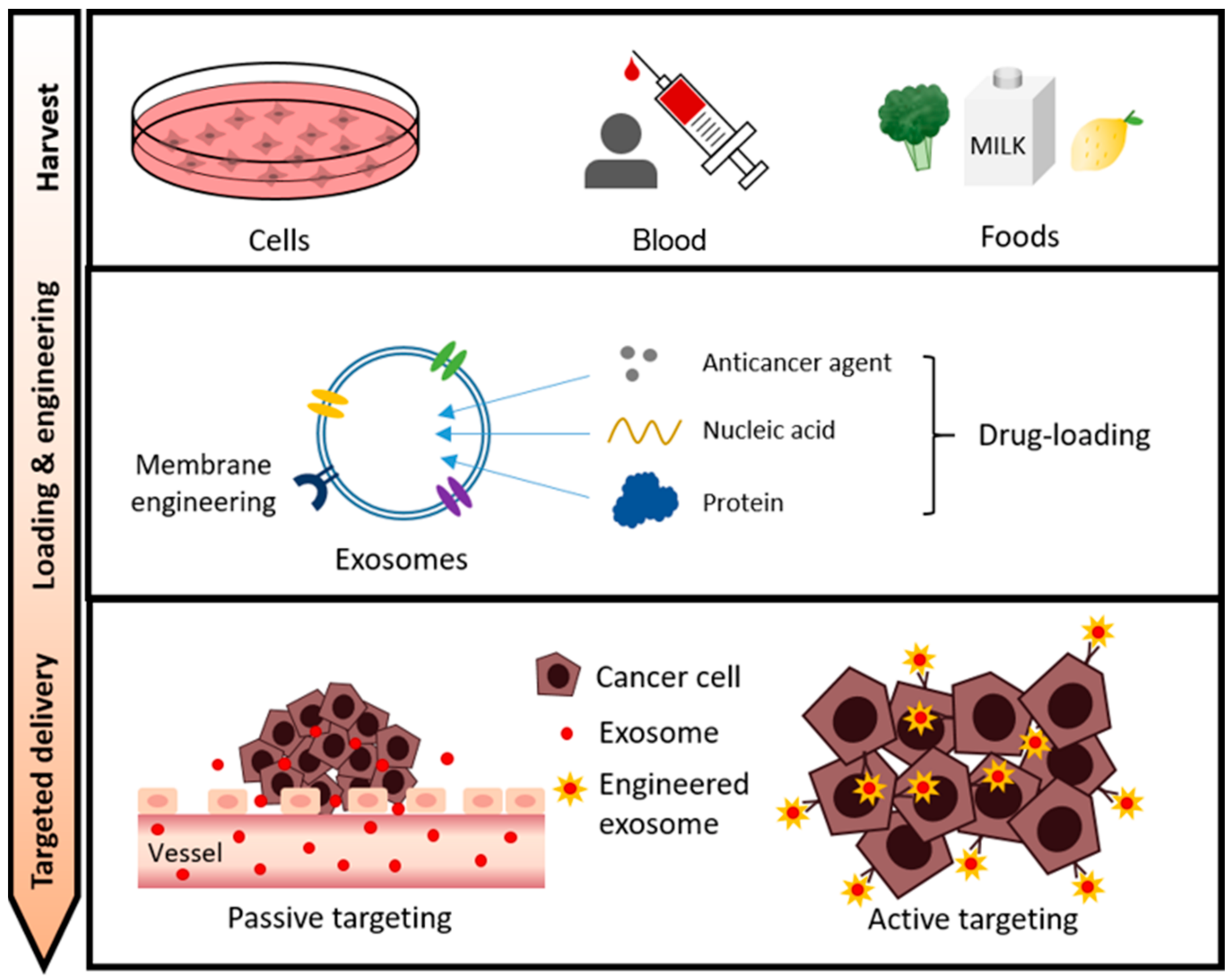
2. Main Types of Exosomes in Drug Delivery Systems
2.1. Cell-Secreted Exosomes
2.1.1. Human Embryonic Kidney Cell (HEK)-Derived Exosomes
2.1.2. Cancer Cell-Derived Exosomes
2.1.3. Immune Cell-Derived Exosomes
2.1.4. Stem Cell-Derived Exosomes
2.2. Blood (Red Blood Cell)-Derived Exosomes
2.3. Food-Derived Exosomes
2.3.1. Milk-Derived Exosomes
2.3.2. Edible Plant-Derived EXOSOMES
3. Exosome Cargo Loading for Cancer Therapy
3.1. Anti-Cancer Drugs
| Cargo | Origin of Exosomes | Target Cancer Type | Loading Method | Administration Route | Outcome | References | |
|---|---|---|---|---|---|---|---|
| Doxorubicin | Monocyte | Colon carcinoma | Incubation | I.V. | Suppression of tumor growth | [94] | |
| Breast cancer cell Ovarian cancer cell | Breast cancer Ovarian cancer | Electroporation | I.P. | Suppression of tumor growth Reducing toxicity of drug | [96] | ||
| MSC | Osteosarcoma myocardial | Incubation | N/A | Reducing toxicity of drug Enhanced the cellular uptake rate | [97] | ||
| Paclitaxel | MSC | Pancreatic adenocarcinoma | Incubation | N/A | Improved anticancer effect | [102] | |
| Prostate cancer cell | Prostate cancer | Incubation | N/A | Overcame drug resistance Enhanced cytotoxicity on cancer cells | [103] | ||
| Macrophage | Pulmonary metastasis | Incubation Electroporation Sonication | I.N. | Overcame MDR in cancer cells Enhanced the cellular uptake rate | [104] | ||
| Bovine milk | Lung cancer | Incubation | P.O. | Suppression of tumor growth Lower systemic toxicity | [80] | ||
| Brain cancer cell | Glioblastoma-astrocytoma cancer | Incubation Sonication | N/A | Enhanced cytotoxicity on cancer cells Passed through the BBB | [98,105] | ||
| Nucleic acid | miRNA-122 | AMSC | Hepatocellular carcinoma | Transfection | I.T. | Improved the chemosensitivity Improved anticancer effect | [106] |
| miRNA-21 | HEK293T cell | Brain tumor | Electroporation | I.V. | Suppression of tumor growth Passed through the BBB | [107] | |
| siRNA | Breast cancer cell | Breast adenocarcinoma | Electroporation | I.V. | Suppression of tumor growth Improved effect of tumor targeting | [108] | |
| Breast cancer cell | Breast cancer Lung metastasis | Incubation | I.V. | Enhanced the cellular uptake rate Reduced migration and metastasis of cancer cells | [109] | ||
| Circular RNA | Hepatocellular cancer cell | Hepatocellular cancer | Pre-overexpression | I.T. | Suppression of tumor growth Promoted tumor cell apoptosis | [110] | |
| mRNA | Red blood cell | Breast cancer Leukemia | Electroporation | I.T. | Suppression of tumor growth Reduced drug cytotoxicity | [43] | |
| Protein | Surviving-T34A | Melanoma cell | Pancreatic adenocarcinoma | Transfection | N/A | Induced apoptosis in adenocarcinoma cells Enhanced drug sensitivity | [111] |
| TRP2 | Serum | Any type of cancer | Saponin/electroporation | S.C. | Accumulation in lymph nodes for immunotherapy | [112] | |
| SIRPα | HEK293T cell | Colon carcinoma | Transfection | I.V. | Enhanced phagocytosis of tumor cells Suppression of tumor growth | [113] | |
| hMUC1 | Colon cancer cell Breast cancer cell | Colon carcinoma | Transfection | I.D. | Proliferation and activation of immune cells Suppression of tumor growth | [114] | |
3.2. Nucleic Acids
3.3. Proteins
4. Exosome Membrane Modifications for Specific Targeting
4.1. Passive Tumor Targeting
4.2. Active Tumor Targeting
5. Challenges and Perspectives
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell. Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Hong, Y.; Cho, E.; Kim, G.B.; Kim, I.S. Extracellular vesicles as a platform for membrane-associated therapeutic protein delivery. J. Extracell. Vesicles 2018, 7, 1440131. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Zhao, J.; Zhang, L.; Wei, F.; Lian, Y.; Wu, Y.; Gong, Z.; Zhang, S.; Zhou, J.; Cao, K.; et al. Role of tumor microenvironment in tumorigenesis. J. Cancer 2017, 8, 761–773. [Google Scholar] [CrossRef]
- Wang, X.; He, L.; Huang, X.; Zhang, S.; Cao, W.; Che, F.; Zhu, Y.; Dai, J. Recent Progress of Exosomes in Multiple Myeloma: Pathogenesis, Diagnosis, Prognosis and Therapeutic Strategies. Cancers 2021, 13, 1635. [Google Scholar] [CrossRef] [PubMed]
- Campanella, C.; Caruso Bavisotto, C.; Logozzi, M.; Marino Gammazza, A.; Mizzoni, D.; Cappello, F.; Fais, S. On the Choice of the Extracellular Vesicles for Therapeutic Purposes. Int. J. Mol. Sci. 2019, 20, 236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Su, C. Design strategies and application progress of therapeutic exosomes. Theranostics 2019, 9, 1015–1028. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.Z.; Ruan, J.S.; Jiang, Z.S.; Wang, L.; Wang, S.M. Extracellular Vesicles: A New Perspective in Tumor Therapy. Biomed. Res. Int. 2018, 2018, 2687954. [Google Scholar] [CrossRef] [Green Version]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3, 24641. [Google Scholar] [CrossRef] [Green Version]
- Purushothaman, A.; Bandari, S.K.; Liu, J.; Mobley, J.A.; Brown, E.E.; Sanderson, R.D. Fibronectin on the Surface of Myeloma Cell-derived Exosomes Mediates Exosome-Cell Interactions. J. Biol. Chem. 2016, 291, 1652–1663. [Google Scholar] [CrossRef] [Green Version]
- Soltani, F.; Parhiz, H.; Mokhtarzadeh, A.; Ramezani, M. Synthetic and Biological Vesicular Nano-Carriers Designed for Gene Delivery. Curr. Pharm. Des. 2015, 21, 6214–6235. [Google Scholar] [CrossRef]
- Gutierrez-Vazquez, C.; Villarroya-Beltri, C.; Mittelbrunn, M.; Sanchez-Madrid, F. Transfer of extracellular vesicles during immune cell-cell interactions. Immunol. Rev. 2013, 251, 125–142. [Google Scholar] [CrossRef] [Green Version]
- Haney, M.J.; Klyachko, N.L.; Zhao, Y.; Gupta, R.; Plotnikova, E.G.; He, Z.; Patel, T.; Piroyan, A.; Sokolsky, M.; Kabanov, A.V.; et al. Exosomes as drug delivery vehicles for Parkinson’s disease therapy. J. Control. Release 2015, 207, 18–30. [Google Scholar] [CrossRef] [Green Version]
- Whiteside, T.L. Immune modulation of T-cell and NK (natural killer) cell activities by TEXs (tumour-derived exosomes). Biochem. Soc. Trans. 2013, 41, 245–251. [Google Scholar] [CrossRef]
- Kawikova, I.; Askenase, P.W. Diagnostic and therapeutic potentials of exosomes in CNS diseases. Brain Res. 2015, 1617, 63–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Tsibouklis, J.; Weng, T.; Zhang, B.; Yin, G.; Feng, G.; Cui, Y.; Savina, I.N.; Mikhalovska, L.I.; Sandeman, S.R.; et al. Nano carriers for drug transport across the blood-brain barrier. J. Drug Target. 2017, 25, 17–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, D.; Zhuang, X.; Xiang, X.; Liu, Y.; Zhang, S.; Liu, C.; Barnes, S.; Grizzle, W.; Miller, D.; Zhang, H.G. A novel nanoparticle drug delivery system: The anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Mol. Ther. 2010, 18, 1606–1614. [Google Scholar] [CrossRef] [PubMed]
- Prada, I.; Meldolesi, J. Binding and Fusion of Extracellular Vesicles to the Plasma Membrane of Their Cell Targets. Int. J. Mol. Sci. 2016, 17, 1296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Andaloussi, S.; Lakhal, S.; Mager, I.; Wood, M.J. Exosomes for targeted siRNA delivery across biological barriers. Adv. Drug Deliv. Rev. 2013, 65, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Valapala, M.; Vishwanatha, J.K. Lipid raft endocytosis and exosomal transport facilitate extracellular trafficking of annexin A2. J. Biol. Chem. 2011, 286, 30911–30925. [Google Scholar] [CrossRef] [Green Version]
- Das, C.K.; Jena, B.C.; Banerjee, I.; Das, S.; Parekh, A.; Bhutia, S.K.; Mandal, M. Exosome as a Novel Shuttle for Delivery of Therapeutics across Biological Barriers. Mol. Pharm. 2019, 16, 24–40. [Google Scholar] [CrossRef]
- Kim, H.; Wang, S.Y.; Kwak, G.; Yang, Y.; Kwon, I.C.; Kim, S.H. Exosome-Guided Phenotypic Switch of M1 to M2 Macrophages for Cutaneous Wound Healing. Adv. Sci. 2019, 6, 1900513. [Google Scholar] [CrossRef] [Green Version]
- Reif, S.; Elbaum-Shiff, Y.; Koroukhov, N.; Shilo, I.; Musseri, M.; Golan-Gerstl, R. Cow and Human Milk-Derived Exosomes Ameliorate Colitis in DSS Murine Model. Nutrients 2020, 12, 2589. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Huang, L.; Li, Y.; Zhang, X.; Gu, J.; Yan, Y.; Xu, X.; Wang, M.; Qian, H.; Xu, W. Exosomes derived from human bone marrow mesenchymal stem cells promote tumor growth in vivo. Cancer Lett. 2012, 315, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Alqahtani, M.S.; Kazi, M.; Alsenaidy, M.A.; Ahmad, M.Z. Advances in Oral Drug Delivery. Front. Pharmacol. 2021, 12, 618411. [Google Scholar] [CrossRef] [PubMed]
- Ismail, R.; Bocsik, A.; Katona, G.; Grof, I.; Deli, M.A.; Csoka, I. Encapsulation in Polymeric Nanoparticles Enhances the Enzymatic Stability and the Permeability of the GLP-1 Analog, Liraglutide, Across a Culture Model of Intestinal Permeability. Pharmaceutics 2019, 11, 599. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Qin, X.; Kong, F.; Chen, P.; Pan, G. Improving cellular uptake of therapeutic entities through interaction with components of cell membrane. Drug Deliv. 2019, 26, 328–342. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.; Chen, L.; Ye, S.; Kang, Y.; Liu, J.; Zeng, S.; Yu, L. Research and development of drug delivery systems based on drug transporter and nano-formulation. Asian J. Pharm. Sci. 2020, 15, 220–236. [Google Scholar] [CrossRef] [PubMed]
- Charoenviriyakul, C.; Takahashi, Y.; Morishita, M.; Matsumoto, A.; Nishikawa, M.; Takakura, Y. Cell type-specific and common characteristics of exosomes derived from mouse cell lines: Yield, physicochemical properties, and pharmacokinetics. Eur. J. Pharm. Sci. 2017, 96, 316–322. [Google Scholar] [CrossRef]
- Vader, P.; Mol, E.A.; Pasterkamp, G.; Schiffelers, R.M. Extracellular vesicles for drug delivery. Adv. Drug Deliv. Rev. 2016, 106, 148–156. [Google Scholar] [CrossRef]
- Zhu, X.; Badawi, M.; Pomeroy, S.; Sutaria, D.S.; Xie, Z.; Baek, A.; Jiang, J.; Elgamal, O.A.; Mo, X.; Perle, K.; et al. Comprehensive toxicity and immunogenicity studies reveal minimal effects in mice following sustained dosing of extracellular vesicles derived from HEK293T cells. J. Extracell. Vesicles 2017, 6, 1324730. [Google Scholar] [CrossRef]
- Li, J.; Chen, X.; Yi, J.; Liu, Y.; Li, D.; Wang, J.; Hou, D.; Jiang, X.; Zhang, J.; Wang, J.; et al. Identification and Characterization of 293T Cell-Derived Exosomes by Profiling the Protein, mRNA and MicroRNA Components. PLoS ONE 2016, 11, e0163043. [Google Scholar] [CrossRef]
- Shao, J.; Zaro, J.; Shen, Y. Advances in Exosome-Based Drug Delivery and Tumor Targeting: From Tissue Distribution to Intracellular Fate. Int. J. Nanomed. 2020, 15, 9355–9371. [Google Scholar] [CrossRef]
- Smyth, T.; Kullberg, M.; Malik, N.; Smith-Jones, P.; Graner, M.W.; Anchordoquy, T.J. Biodistribution and delivery efficiency of unmodified tumor-derived exosomes. J. Control. Release 2015, 199, 145–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiao, L.; Hu, S.; Huang, K.; Su, T.; Li, Z.; Vandergriff, A.; Cores, J.; Dinh, P.U.; Allen, T.; Shen, D.; et al. Tumor cell-derived exosomes home to their cells of origin and can be used as Trojan horses to deliver cancer drugs. Theranostics 2020, 10, 3474–3487. [Google Scholar] [CrossRef] [PubMed]
- Munich, S.; Sobo-Vujanovic, A.; Buchser, W.J.; Beer-Stolz, D.; Vujanovic, N.L. Dendritic cell exosomes directly kill tumor cells and activate natural killer cells via TNF superfamily ligands. Oncoimmunology 2012, 1, 1074–1083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pitt, J.M.; Andre, F.; Amigorena, S.; Soria, J.C.; Eggermont, A.; Kroemer, G.; Zitvogel, L. Dendritic cell-derived exosomes for cancer therapy. J. Clin. Investig. 2016, 126, 1224–1232. [Google Scholar] [CrossRef] [PubMed]
- Viaud, S.; Terme, M.; Flament, C.; Taieb, J.; Andre, F.; Novault, S.; Escudier, B.; Robert, C.; Caillat-Zucman, S.; Tursz, T.; et al. Dendritic cell-derived exosomes promote natural killer cell activation and proliferation: A role for NKG2D ligands and IL-15Ralpha. PLoS ONE 2009, 4, e4942. [Google Scholar] [CrossRef]
- Simhadri, V.R.; Reiners, K.S.; Hansen, H.P.; Topolar, D.; Simhadri, V.L.; Nohroudi, K.; Kufer, T.A.; Engert, A.; Pogge von Strandmann, E. Dendritic cells release HLA-B-associated transcript-3 positive exosomes to regulate natural killer function. PLoS ONE 2008, 3, e3377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andre, F.; Escudier, B.; Angevin, E.; Tursz, T.; Zitvogel, L. Exosomes for cancer immunotherapy. Ann. Oncol. 2004, 15 (Suppl. 4), iv141–iv144. [Google Scholar] [CrossRef]
- Baek, G.; Choi, H.; Kim, Y.; Lee, H.C.; Choi, C. Mesenchymal Stem Cell-Derived Extracellular Vesicles as Therapeutics and as a Drug Delivery Platform. Stem Cells Transl. Med. 2019, 8, 880–886. [Google Scholar] [CrossRef] [Green Version]
- Yeo, R.W.; Lai, R.C.; Zhang, B.; Tan, S.S.; Yin, Y.; Teh, B.J.; Lim, S.K. Mesenchymal stem cell: An efficient mass producer of exosomes for drug delivery. Adv. Drug Deliv. Rev. 2013, 65, 336–341. [Google Scholar] [CrossRef]
- Usman, W.M.; Pham, T.C.; Kwok, Y.Y.; Vu, L.T.; Ma, V.; Peng, B.; Chan, Y.S.; Wei, L.; Chin, S.M.; Azad, A.; et al. Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat. Commun. 2018, 9, 2359. [Google Scholar] [CrossRef] [PubMed]
- Gangadaran, P.; Ahn, B.C. Extracellular Vesicle- and Extracellular Vesicle Mimetics-Based Drug Delivery Systems: New Perspectives, Challenges, and Clinical Developments. Pharmaceutics 2020, 12, 442. [Google Scholar] [CrossRef] [PubMed]
- Izumi, H.; Kosaka, N.; Shimizu, T.; Sekine, K.; Ochiya, T.; Takase, M. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J. Dairy Sci. 2012, 95, 4831–4841. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Du, X.; Li, J.; Lonnerdal, B. Human milk exosomes and their microRNAs survive digestion in vitro and are taken up by human intestinal cells. Mol. Nutr. Food Res. 2017, 61, 1700082. [Google Scholar] [CrossRef]
- Adriano, B.; Cotto, N.M.; Chauhan, N.; Jaggi, M.; Chauhan, S.C.; Yallapu, M.M. Milk exosomes: Nature’s abundant nanoplatform for theranostic applications. Bioact. Mater. 2021, 6, 2479–2490. [Google Scholar] [CrossRef]
- Shandilya, S.; Rani, P.; Onteru, S.K.; Singh, D. Small Interfering RNA in Milk Exosomes Is Resistant to Digestion and Crosses the Intestinal Barrier In Vitro. J. Agric. Food Chem. 2017, 65, 9506–9513. [Google Scholar] [CrossRef]
- Zhang, M.; Xiao, B.; Wang, H.; Han, M.K.; Zhang, Z.; Viennois, E.; Xu, C.; Merlin, D. Edible Ginger-derived Nano-lipids Loaded with Doxorubicin as a Novel Drug-delivery Approach for Colon Cancer Therapy. Mol. Ther. 2016, 24, 1783–1796. [Google Scholar] [CrossRef] [Green Version]
- Ju, S.; Mu, J.; Dokland, T.; Zhuang, X.; Wang, Q.; Jiang, H.; Xiang, X.; Deng, Z.B.; Wang, B.; Zhang, L.; et al. Grape exosome-like nanoparticles induce intestinal stem cells and protect mice from DSS-induced colitis. Mol. Ther. 2013, 21, 1345–1357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dad, H.A.; Gu, T.W.; Zhu, A.Q.; Huang, L.Q.; Peng, L.H. Plant Exosome-like Nanovesicles: Emerging Therapeutics and Drug Delivery Nanoplatforms. Mol. Ther. 2021, 29, 13–31. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Zaborowski, M.P.; Balaj, L.; Breakefield, X.O.; Lai, C.P. Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. Bioscience 2015, 65, 783–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lobb, R.J.; Becker, M.; Wen, S.W.; Wong, C.S.; Wiegmans, A.P.; Leimgruber, A.; Moller, A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 2015, 4, 27031. [Google Scholar] [CrossRef]
- Li, P.; Kaslan, M.; Lee, S.H.; Yao, J.; Gao, Z. Progress in Exosome Isolation Techniques. Theranostics 2017, 7, 789–804. [Google Scholar] [CrossRef] [PubMed]
- Jurgielewicz, B.J.; Yao, Y.; Stice, S.L. Kinetics and Specificity of HEK293T Extracellular Vesicle Uptake using Imaging Flow Cytometry. Nanoscale Res. Lett. 2020, 15, 170. [Google Scholar] [CrossRef] [PubMed]
- Kanada, M.; Bachmann, M.H.; Hardy, J.W.; Frimannson, D.O.; Bronsart, L.; Wang, A.; Sylvester, M.D.; Schmidt, T.L.; Kaspar, R.L.; Butte, M.J.; et al. Differential fates of biomolecules delivered to target cells via extracellular vesicles. Proc. Natl. Acad. Sci. USA 2015, 112, E1433–E1442. [Google Scholar] [CrossRef] [Green Version]
- Vilanova-Perez, T.; Jones, C.; Balint, S.; Dragovic, R.; Dustin, M.L.; Yeste, M.; Coward, K. Exosomes derived from HEK293T cells interact in an efficient and noninvasive manner with mammalian sperm in vitro. Nanomedicine 2020, 15, 1965–1980. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.B.; Nam, G.H.; Hong, Y.; Woo, J.; Cho, Y.; Kwon, I.C.; Yang, Y.; Kim, I.S. Xenogenization of tumor cells by fusogenic exosomes in tumor microenvironment ignites and propagates antitumor immunity. Sci. Adv. 2020, 6, eaaz2083. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Nam, G.-H.; Koh, E.; Jeon, S.; Kim, G.B.; Jeong, C.; Kim, D.-H.; Yang, Y.; Kim, I.-S. Exosome as a Vehicle for Delivery of Membrane Protein Therapeutics, PH20, for Enhanced Tumor Penetration and Antitumor Efficacy. Adv. Funct. Mater. 2018, 28, 1703074. [Google Scholar] [CrossRef]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell. Biol. 2010, 12, 19–30. [Google Scholar] [CrossRef] [Green Version]
- Hermans, I.F.; Ritchie, D.S.; Yang, J.; Roberts, J.M.; Ronchese, F. CD8+ T cell-dependent elimination of dendritic cells in vivo limits the induction of antitumor immunity. J. Immunol. 2000, 164, 3095–3101. [Google Scholar] [CrossRef] [Green Version]
- Quah, B.J.; O’Neill, H.C. The immunogenicity of dendritic cell-derived exosomes. Blood Cells Mol. Dis. 2005, 35, 94–110. [Google Scholar] [CrossRef] [PubMed]
- Giordano, A.; Galderisi, U.; Marino, I.R. From the laboratory bench to the patient’s bedside: An update on clinical trials with mesenchymal stem cells. J. Cell. Physiol. 2007, 211, 27–35. [Google Scholar] [CrossRef]
- in ’t Anker, P.S.; Noort, W.A.; Scherjon, S.A.; Kleijburg-van der Keur, C.; Kruisselbrink, A.B.; van Bezooijen, R.L.; Beekhuizen, W.; Willemze, R.; Kanhai, H.H.; Fibbe, W.E. Mesenchymal stem cells in human second-trimester bone marrow, liver, lung, and spleen exhibit a similar immunophenotype but a heterogeneous multilineage differentiation potential. Haematologica 2003, 88, 845–852. [Google Scholar] [PubMed]
- Mendt, M.; Kamerkar, S.; Sugimoto, H.; McAndrews, K.M.; Wu, C.C.; Gagea, M.; Yang, S.; Blanko, E.V.R.; Peng, Q.; Ma, X.; et al. Generation and testing of clinical-grade exosomes for pancreatic cancer. JCI Insight 2018, 3, e99263. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Xu, K.; Zheng, X.; Chen, T.; Wang, J.; Song, Y.; Shao, Y.; Zheng, S. Application of exosomes as liquid biopsy in clinical diagnosis. Signal. Transduct. Target. Ther. 2020, 5, 144. [Google Scholar] [CrossRef]
- Yanez-Mo, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borras, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [Green Version]
- Keller, S.; Ridinger, J.; Rupp, A.K.; Janssen, J.W.; Altevogt, P. Body fluid derived exosomes as a novel template for clinical diagnostics. J. Transl. Med. 2011, 9, 86. [Google Scholar] [CrossRef] [Green Version]
- Sheller-Miller, S.; Menon, R. Isolation and characterization of human amniotic fluid-derived exosomes. Methods Enzymol. 2020, 645, 181–194. [Google Scholar] [CrossRef]
- Gonzales, P.A.; Zhou, H.; Pisitkun, T.; Wang, N.S.; Star, R.A.; Knepper, M.A.; Yuen, P.S. Isolation and purification of exosomes in urine. Methods Mol. Biol. 2010, 641, 89–99. [Google Scholar] [CrossRef]
- Baranyai, T.; Herczeg, K.; Onodi, Z.; Voszka, I.; Modos, K.; Marton, N.; Nagy, G.; Mager, I.; Wood, M.J.; El Andaloussi, S.; et al. Isolation of Exosomes from Blood Plasma: Qualitative and Quantitative Comparison of Ultracentrifugation and Size Exclusion Chromatography Methods. PLoS ONE 2015, 10, e0145686. [Google Scholar] [CrossRef] [Green Version]
- Cheshmi, B.; Cheshomi, H. Salivary exosomes: Properties, medical applications, and isolation methods. Mol. Biol. Rep. 2020, 47, 6295–6307. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Kundrat, L.; Pishesha, N.; Bilate, A.; Theile, C.; Maruyama, T.; Dougan, S.K.; Ploegh, H.L.; Lodish, H.F. Engineered red blood cells as carriers for systemic delivery of a wide array of functional probes. Proc. Natl. Acad. Sci. USA 2014, 111, 10131–10136. [Google Scholar] [CrossRef] [Green Version]
- Munagala, R.; Aqil, F.; Jeyabalan, J.; Gupta, R.C. Bovine milk-derived exosomes for drug delivery. Cancer Lett. 2016, 371, 48–61. [Google Scholar] [CrossRef] [Green Version]
- Haug, A.; Hostmark, A.T.; Harstad, O.M. Bovine milk in human nutrition—A review. Lipids Health Dis. 2007, 6, 25. [Google Scholar] [CrossRef] [Green Version]
- Torregrosa Paredes, P.; Gutzeit, C.; Johansson, S.; Admyre, C.; Stenius, F.; Alm, J.; Scheynius, A.; Gabrielsson, S. Differences in exosome populations in human breast milk in relation to allergic sensitization and lifestyle. Allergy 2014, 69, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Milk: Production per Cow by Year, US. Available online: https://www.nass.usda.gov/Charts_and_Maps/Milk_Production_and_Milk_Cows/cowrates.php (accessed on 23 February 2021).
- Agrawal, A.K.; Aqil, F.; Jeyabalan, J.; Spencer, W.A.; Beck, J.; Gachuki, B.W.; Alhakeem, S.S.; Oben, K.; Munagala, R.; Bondada, S.; et al. Milk-derived exosomes for oral delivery of paclitaxel. Nanomedicine 2017, 13, 1627–1636. [Google Scholar] [CrossRef] [PubMed]
- Martins, J.P.; Kennedy, P.J.; Santos, H.A.; Barrias, C.; Sarmento, B. A comprehensive review of the neonatal Fc receptor and its application in drug delivery. Pharmacol. Ther. 2016, 161, 22–39. [Google Scholar] [CrossRef] [PubMed]
- Rodewald, R. pH-dependent binding of immunoglobulins to intestinal cells of the neonatal rat. J. Cell. Biol. 1976, 71, 666–669. [Google Scholar] [CrossRef] [Green Version]
- Sockolosky, J.T.; Szoka, F.C. The neonatal Fc receptor, FcRn, as a target for drug delivery and therapy. Adv. Drug Deliv. Rev. 2015, 91, 109–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warren, M.R.; Zhang, C.; Vedadghavami, A.; Bokvist, K.; Dhal, P.K.; Bajpayee, A.G. Milk exosomes with enhanced mucus penetrability for oral delivery of siRNA. Biomater. Sci. 2020, 9, 4260–4277. [Google Scholar] [CrossRef] [PubMed]
- Mu, J.; Zhuang, X.; Wang, Q.; Jiang, H.; Deng, Z.B.; Wang, B.; Zhang, L.; Kakar, S.; Jun, Y.; Miller, D.; et al. Interspecies communication between plant and mouse gut host cells through edible plant derived exosome-like nanoparticles. Mol. Nutr. Food Res. 2014, 58, 1561–1573. [Google Scholar] [CrossRef]
- Wang, B.; Zhuang, X.; Deng, Z.B.; Jiang, H.; Mu, J.; Wang, Q.; Xiang, X.; Guo, H.; Zhang, L.; Dryden, G.; et al. Targeted drug delivery to intestinal macrophages by bioactive nanovesicles released from grapefruit. Mol. Ther. 2014, 22, 522–534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giallombardo, M.; Taverna, S.; Alessandro, R.; Hong, D.; Rolfo, C. Exosome-mediated drug resistance in cancer: The near future is here. Ther. Adv. Med. Oncol. 2016, 8, 320–322. [Google Scholar] [CrossRef] [Green Version]
- Song, H.; Liu, B.; Dong, B.; Xu, J.; Zhou, H.; Na, S.; Liu, Y.; Pan, Y.; Chen, F.; Li, L.; et al. Exosome-Based Delivery of Natural Products in Cancer Therapy. Front. Cell. Dev. Biol. 2021, 9, 650426. [Google Scholar] [CrossRef]
- Wang, J.; Zheng, Y.; Zhao, M. Exosome-Based Cancer Therapy: Implication for Targeting Cancer Stem Cells. Front. Pharmacol. 2016, 7, 533. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.D.; Jiang, L.H.; Hou, J.C.; Zhong, S.L.; Zhu, L.P.; Wang, D.D.; Zhou, S.Y.; Yang, S.J.; Wang, J.Y.; Zhang, Q.; et al. Exosome: A novel mediator in drug resistance of cancer cells. Epigenomics 2018, 10, 1499–1509. [Google Scholar] [CrossRef]
- Zhao, X.; Wu, D.; Ma, X.; Wang, J.; Hou, W.; Zhang, W. Exosomes as drug carriers for cancer therapy and challenges regarding exosome uptake. Biomed. Pharmacother. 2020, 128, 110237. [Google Scholar] [CrossRef] [PubMed]
- Kanwal, U.; Irfan Bukhari, N.; Ovais, M.; Abass, N.; Hussain, K.; Raza, A. Advances in nano-delivery systems for doxorubicin: An updated insight. J. Drug Target. 2018, 26, 296–310. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Woodle, M.C.; Mixson, A.J. Advances in delivery systems for doxorubicin. J. Nanomed. Nanotechnol. 2018, 9, 519. [Google Scholar] [CrossRef]
- Jang, S.C.; Kim, O.Y.; Yoon, C.M.; Choi, D.S.; Roh, T.Y.; Park, J.; Nilsson, J.; Lotvall, J.; Kim, Y.K.; Gho, Y.S. Bioinspired exosome-mimetic nanovesicles for targeted delivery of chemotherapeutics to malignant tumors. ACS Nano 2013, 7, 7698–7710. [Google Scholar] [CrossRef] [PubMed]
- Smyth, T.J.; Redzic, J.S.; Graner, M.W.; Anchordoquy, T.J. Examination of the specificity of tumor cell derived exosomes with tumor cells in vitro. Biochim. Biophys. Acta 2014, 1838, 2954–2965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hadla, M.; Palazzolo, S.; Corona, G.; Caligiuri, I.; Canzonieri, V.; Toffoli, G.; Rizzolio, F. Exosomes increase the therapeutic index of doxorubicin in breast and ovarian cancer mouse models. Nanomedicine 2016, 11, 2431–2441. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Chen, J.; Wang, S.; Fu, F.; Zhu, X.; Wu, C.; Liu, Z.; Zhong, G.; Lin, J. A Nanodrug Consisting Of Doxorubicin And Exosome Derived From Mesenchymal Stem Cells For Osteosarcoma Treatment In Vitro. Int. J. Nanomed. 2019, 14, 8603–8610. [Google Scholar] [CrossRef] [Green Version]
- Salarpour, S.; Forootanfar, H.; Pournamdari, M.; Ahmadi-Zeidabadi, M.; Esmaeeli, M.; Pardakhty, A. Paclitaxel incorporated exosomes derived from glioblastoma cells: Comparative study of two loading techniques. Daru 2019, 27, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Mu, Q.; Jeon, M.; Hsiao, M.H.; Patton, V.K.; Wang, K.; Press, O.W.; Zhang, M. Stable and efficient Paclitaxel nanoparticles for targeted glioblastoma therapy. Adv. Healthc. Mater. 2015, 4, 1236–1245. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Wang, H.; Huang, Q.; Peng, C.; Yao, L.; Chen, H.; Qiu, Z.; Wu, Y.; Wang, L.; Chen, W. Exosomes from M1-Polarized Macrophages Enhance Paclitaxel Antitumor Activity by Activating Macrophages-Mediated Inflammation. Theranostics 2019, 9, 1714–1727. [Google Scholar] [CrossRef]
- Xin, H.; Sha, X.; Jiang, X.; Zhang, W.; Chen, L.; Fang, X. Anti-glioblastoma efficacy and safety of paclitaxel-loading Angiopep-conjugated dual targeting PEG-PCL nanoparticles. Biomaterials 2012, 33, 8167–8176. [Google Scholar] [CrossRef]
- Pascucci, L.; Cocce, V.; Bonomi, A.; Ami, D.; Ceccarelli, P.; Ciusani, E.; Vigano, L.; Locatelli, A.; Sisto, F.; Doglia, S.M.; et al. Paclitaxel is incorporated by mesenchymal stromal cells and released in exosomes that inhibit in vitro tumor growth: A new approach for drug delivery. J. Control. Release 2014, 192, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Saari, H.; Lazaro-Ibanez, E.; Viitala, T.; Vuorimaa-Laukkanen, E.; Siljander, P.; Yliperttula, M. Microvesicle- and exosome-mediated drug delivery enhances the cytotoxicity of Paclitaxel in autologous prostate cancer cells. J. Control. Release 2015, 220, 727–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.S.; Haney, M.J.; Zhao, Y.; Mahajan, V.; Deygen, I.; Klyachko, N.L.; Inskoe, E.; Piroyan, A.; Sokolsky, M.; Okolie, O.; et al. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine 2016, 12, 655–664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gan, H.K.; Rosenthal, M.A.; Cher, L.; Dally, M.; Drummond, K.; Murphy, M.; Thursfield, V. Management of glioblastoma in Victoria, Australia (2006–2008). J. Clin. Neurosci. 2015, 22, 1462–1466. [Google Scholar] [CrossRef]
- Lou, G.; Song, X.; Yang, F.; Wu, S.; Wang, J.; Chen, Z.; Liu, Y. Exosomes derived from miR-122-modified adipose tissue-derived MSCs increase chemosensitivity of hepatocellular carcinoma. J. Hematol. Oncol. 2015, 8, 122. [Google Scholar] [CrossRef] [Green Version]
- Kim, G.; Kim, M.; Lee, Y.; Byun, J.W.; Hwang, D.W.; Lee, M. Systemic delivery of microRNA-21 antisense oligonucleotides to the brain using T7-peptide decorated exosomes. J. Control. Release 2020, 317, 273–281. [Google Scholar] [CrossRef]
- Yang, Z.; Xie, J.; Zhu, J.; Kang, C.; Chiang, C.; Wang, X.; Wang, X.; Kuang, T.; Chen, F.; Chen, Z.; et al. Functional exosome-mimic for delivery of siRNA to cancer: In vitro and in vivo evaluation. J. Control. Release 2016, 243, 160–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Gu, C.; Gan, Y.; Shao, L.; Chen, H.; Zhu, H. Exosome-mediated siRNA delivery to suppress postoperative breast cancer metastasis. J. Control. Release 2020, 318, 1–15. [Google Scholar] [CrossRef]
- Chen, W.; Quan, Y.; Fan, S.; Wang, H.; Liang, J.; Huang, L.; Chen, L.; Liu, Q.; He, P.; Ye, Y. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 2020, 475, 119–128. [Google Scholar] [CrossRef]
- Aspe, J.R.; Diaz Osterman, C.J.; Jutzy, J.M.; Deshields, S.; Whang, S.; Wall, N.R. Enhancement of Gemcitabine sensitivity in pancreatic adenocarcinoma by novel exosome-mediated delivery of the Survivin-T34A mutant. J. Extracell. Vesicles 2014, 3, 23244. [Google Scholar] [CrossRef]
- Park, O.; Choi, E.S.; Yu, G.; Kim, J.Y.; Kang, Y.Y.; Jung, H.; Mok, H. Efficient Delivery of Tyrosinase Related Protein-2 (TRP2) Peptides to Lymph Nodes using Serum-Derived Exosomes. Macromol. Biosci. 2018, 18, e1800301. [Google Scholar] [CrossRef]
- Koh, E.; Lee, E.J.; Nam, G.H.; Hong, Y.; Cho, E.; Yang, Y.; Kim, I.S. Exosome-SIRPalpha, a CD47 blockade increases cancer cell phagocytosis. Biomaterials 2017, 121, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.A.; Yeo, D.J.; Son, H.Y.; Kim, H.W.; Jung, D.S.; Ko, J.K.; Koh, J.S.; Kim, Y.N.; Kim, C.W. Exosomes: A new delivery system for tumor antigens in cancer immunotherapy. Int. J. Cancer 2005, 114, 613–622. [Google Scholar] [CrossRef]
- Carobolante, G.; Mantaj, J.; Ferrari, E.; Vllasaliu, D. Cow Milk and Intestinal Epithelial Cell-derived Extracellular Vesicles as Systems for Enhancing Oral Drug Delivery. Pharmaceutics 2020, 12, 226. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Gao, C.; Kuang, M.; Xu, M.; Wang, B.; Luo, Y.; Teng, L.; Xie, J. Nanoparticles as Drug Delivery Systems of RNAi in Cancer Therapy. Molecules 2021, 26, 2380. [Google Scholar] [CrossRef] [PubMed]
- Nastiuk, K.L.; Krolewski, J.J. Opportunities and challenges in combination gene cancer therapy. Adv. Drug Deliv. Rev. 2016, 98, 35–40. [Google Scholar] [CrossRef] [Green Version]
- Akhtar, S. Non-viral cancer gene therapy: Beyond delivery. Gene Ther. 2006, 13, 739–740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pai, S.I.; Lin, Y.Y.; Macaes, B.; Meneshian, A.; Hung, C.F.; Wu, T.C. Prospects of RNA interference therapy for cancer. Gene Ther. 2006, 13, 464–477. [Google Scholar] [CrossRef] [PubMed]
- Singha, K.; Namgung, R.; Kim, W.J. Polymers in small-interfering RNA delivery. Nucleic Acid. Ther. 2011, 21, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell. Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Johnsen, K.B.; Gudbergsson, J.M.; Duroux, M.; Moos, T.; Andresen, T.L.; Simonsen, J.B. On the use of liposome controls in studies investigating the clinical potential of extracellular vesicle-based drug delivery systems—A commentary. J. Control. Release 2018, 269, 10–14. [Google Scholar] [CrossRef]
- Lu, M.; Zhao, X.; Xing, H.; Xun, Z.; Zhu, S.; Lang, L.; Yang, T.; Cai, C.; Wang, D.; Ding, P. Comparison of exosome-mimicking liposomes with conventional liposomes for intracellular delivery of siRNA. Int. J. Pharm. 2018, 550, 100–113. [Google Scholar] [CrossRef]
- Sabu, C.; Rejo, C.; Kotta, S.; Pramod, K. Bioinspired and biomimetic systems for advanced drug and gene delivery. J. Control. Release 2018, 287, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Zuo, B.; Jing, R.; Gao, X.; Rao, Q.; Liu, Z.; Qi, H.; Guo, H.; Yin, H. Dendritic cell-derived exosomes elicit tumor regression in autochthonous hepatocellular carcinoma mouse models. J. Hepatol. 2017, 67, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Almen, M.S.; Nordstrom, K.J.; Fredriksson, R.; Schioth, H.B. Mapping the human membrane proteome: A majority of the human membrane proteins can be classified according to function and evolutionary origin. BMC Biol. 2009, 7, 50. [Google Scholar] [CrossRef] [Green Version]
- Cho, E.; Nam, G.H.; Hong, Y.; Kim, Y.K.; Kim, D.H.; Yang, Y.; Kim, I.S. Comparison of exosomes and ferritin protein nanocages for the delivery of membrane protein therapeutics. J. Control. Release 2018, 279, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Hood, J.L. Post isolation modification of exosomes for nanomedicine applications. Nanomedicine 2016, 11, 1745–1756. [Google Scholar] [CrossRef] [Green Version]
- Johnsen, K.B.; Gudbergsson, J.M.; Skov, M.N.; Pilgaard, L.; Moos, T.; Duroux, M. A comprehensive overview of exosomes as drug delivery vehicles—Endogenous nanocarriers for targeted cancer therapy. Biochim. Biophys. Acta 2014, 1846, 75–87. [Google Scholar] [CrossRef]
- Yang, Z.; Shi, J.; Xie, J.; Wang, Y.; Sun, J.; Liu, T.; Zhao, Y.; Zhao, X.; Wang, X.; Ma, Y.; et al. Large-scale generation of functional mRNA-encapsulating exosomes via cellular nanoporation. Nat. Biomed. Eng. 2020, 4, 69–83. [Google Scholar] [CrossRef]
- Shi, S.; Li, T.; Wen, X.; Wu, S.Y.; Xiong, C.; Zhao, J.; Lincha, V.R.; Chow, D.S.; Liu, Y.; Sood, A.K.; et al. Copper-64 Labeled PEGylated Exosomes for In Vivo Positron Emission Tomography and Enhanced Tumor Retention. Bioconjug. Chem. 2019, 30, 2675–2683. [Google Scholar] [CrossRef]
- Tian, Y.; Li, S.; Song, J.; Ji, T.; Zhu, M.; Anderson, G.J.; Wei, J.; Nie, G. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials 2014, 35, 2383–2390. [Google Scholar] [CrossRef]
- Bellavia, D.; Raimondo, S.; Calabrese, G.; Forte, S.; Cristaldi, M.; Patinella, A.; Memeo, L.; Manno, M.; Raccosta, S.; Diana, P.; et al. Interleukin 3- receptor targeted exosomes inhibit in vitro and in vivo Chronic Myelogenous Leukemia cell growth. Theranostics 2017, 7, 1333–1345. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Altinoglu, S.; Takeda, Y.S.; Xu, Q. Integrating Protein Engineering and Bioorthogonal Click Conjugation for Extracellular Vesicle Modulation and Intracellular Delivery. PLoS ONE 2015, 10, e0141860. [Google Scholar] [CrossRef] [PubMed]
- Royo, F.; Cossio, U.; Ruiz de Angulo, A.; Llop, J.; Falcon-Perez, J.M. Modification of the glycosylation of extracellular vesicles alters their biodistribution in mice. Nanoscale 2019, 11, 1531–1537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.; Park, H.; Yu, H.S.; Na, K.; Oh, K.T.; Lee, E.S. Dendritic Cell-Targeted pH-Responsive Extracellular Vesicles for Anticancer Vaccination. Pharmaceutics 2019, 11, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, X.; Cheng, Q.; Hou, T.; Han, M.; Smbatyan, G.; Lang, J.E.; Epstein, A.L.; Lenz, H.J.; Zhang, Y. Genetically Engineered Cell-Derived Nanoparticles for Targeted Breast Cancer Immunotherapy. Mol. Ther. 2020, 28, 536–547. [Google Scholar] [CrossRef]
- Si, Y.; Kim, S.; Zhang, E.; Tang, Y.; Jaskula-Sztul, R.; Markert, J.M.; Chen, H.; Zhou, L.; Liu, X.M. Targeted Exosomes for Drug Delivery: Biomanufacturing, Surface Tagging, and Validation. Biotechnol. J. 2020, 15, e1900163. [Google Scholar] [CrossRef]
- Wang, J.; Li, W.; Zhang, L.; Ban, L.; Chen, P.; Du, W.; Feng, X.; Liu, B.F. Chemically Edited Exosomes with Dual Ligand Purified by Microfluidic Device for Active Targeted Drug Delivery to Tumor Cells. ACS Appl. Mater. Interfaces 2017, 9, 27441–27452. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Shi, M.; Liu, X.; Jin, C.; Xing, X.; Qiu, L.; Tan, W. Aptamer-Functionalized Exosomes: Elucidating the Cellular Uptake Mechanism and the Potential for Cancer-Targeted Chemotherapy. Anal. Chem. 2019, 91, 2425–2430. [Google Scholar] [CrossRef]
- Rosenblum, D.; Joshi, N.; Tao, W.; Karp, J.M.; Peer, D. Progress and challenges towards targeted delivery of cancer therapeutics. Nat. Commun. 2018, 9, 1410. [Google Scholar] [CrossRef] [Green Version]
- Chauhan, V.P.; Jain, R.K. Strategies for advancing cancer nanomedicine. Nat. Mater. 2013, 12, 958–962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ojha, T.; Pathak, V.; Shi, Y.; Hennink, W.E.; Moonen, C.T.W.; Storm, G.; Kiessling, F.; Lammers, T. Pharmacological and physical vessel modulation strategies to improve EPR-mediated drug targeting to tumors. Adv. Drug Deliv. Rev. 2017, 119, 44–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, Z.; Zhao, L.; Lu, F.; Gao, X.; Dong, Y.; Zhao, Y.; Wei, M.; Yang, G.; Xing, C.; Liu, L. Mononuclear phagocyte system blockade improves therapeutic exosome delivery to the myocardium. Theranostics 2020, 10, 218–230. [Google Scholar] [CrossRef]
- Smyth, T.; Petrova, K.; Payton, N.M.; Persaud, I.; Redzic, J.S.; Graner, M.W.; Smith-Jones, P.; Anchordoquy, T.J. Surface functionalization of exosomes using click chemistry. Bioconjug. Chem. 2014, 25, 1777–1784. [Google Scholar] [CrossRef] [Green Version]
- Song, S.; Shim, M.K.; Lim, S.; Moon, Y.; Yang, S.; Kim, J.; Hong, Y.; Yoon, H.Y.; Kim, I.S.; Hwang, K.Y.; et al. In Situ One-Step Fluorescence Labeling Strategy of Exosomes via Bioorthogonal Click Chemistry for Real-Time Exosome Tracking In Vitro and In Vivo. Bioconjug. Chem. 2020, 31, 1562–1574. [Google Scholar] [CrossRef]
- Garcia-Manrique, P.; Matos, M.; Gutierrez, G.; Pazos, C.; Blanco-Lopez, M.C. Therapeutic biomaterials based on extracellular vesicles: Classification of bio-engineering and mimetic preparation routes. J. Extracell. Vesicles 2018, 7, 1422676. [Google Scholar] [CrossRef] [Green Version]
- Pucci, M.; Malagolini, N.; Dall’Olio, F. Glycosyltransferase B4GALNT2 as a Predictor of Good Prognosis in Colon Cancer: Lessons from Databases. Int. J. Mol. Sci. 2021, 22, 4331. [Google Scholar] [CrossRef]
- Shin, E.M.; Huynh, V.T.; Neja, S.A.; Liu, C.Y.; Raju, A.; Tan, K.; Tan, N.S.; Gunaratne, J.; Bi, X.; Iyer, L.M.; et al. GREB1: An evolutionarily conserved protein with a glycosyltransferase domain links ERalpha glycosylation and stability to cancer. Sci. Adv. 2021, 7, eabe2470. [Google Scholar] [CrossRef]
- Fuster, M.M.; Esko, J.D. The sweet and sour of cancer: Glycans as novel therapeutic targets. Nat. Rev. Cancer 2005, 5, 526–542. [Google Scholar] [CrossRef] [PubMed]
- Glavey, S.V.; Huynh, D.; Reagan, M.R.; Manier, S.; Moschetta, M.; Kawano, Y.; Roccaro, A.M.; Ghobrial, I.M.; Joshi, L.; O’Dwyer, M.E. The cancer glycome: Carbohydrates as mediators of metastasis. Blood Rev. 2015, 29, 269–279. [Google Scholar] [CrossRef]
- Boedtkjer, E.; Pedersen, S.F. The Acidic Tumor Microenvironment as a Driver of Cancer. Annu. Rev. Physiol. 2020, 82, 103–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.X.; Choi, S.Y.C.; Niu, X.; Kang, N.; Xue, H.; Killam, J.; Wang, Y. Lactic Acid and an Acidic Tumor Microenvironment suppress Anticancer Immunity. Int. J. Mol. Sci. 2020, 21, 8363. [Google Scholar] [CrossRef]
- Qi, H.; Liu, C.; Long, L.; Ren, Y.; Zhang, S.; Chang, X.; Qian, X.; Jia, H.; Zhao, J.; Sun, J.; et al. Blood Exosomes Endowed with Magnetic and Targeting Properties for Cancer Therapy. ACS Nano 2016, 10, 3323–3333. [Google Scholar] [CrossRef]
- Taghikhani, A.; Farzaneh, F.; Sharifzad, F.; Mardpour, S.; Ebrahimi, M.; Hassan, Z.M. Engineered Tumor-Derived Extracellular Vesicles: Potentials in Cancer Immunotherapy. Front. Immunol. 2020, 11, 221. [Google Scholar] [CrossRef]
- Tan, Y.; Luo, X.; Lv, W.; Hu, W.; Zhao, C.; Xiong, M.; Yi, Y.; Wang, D.; Wang, Y.; Wang, H.; et al. Tumor-derived exosomal components: The multifaceted roles and mechanisms in breast cancer metastasis. Cell Death Dis. 2021, 12, 547. [Google Scholar] [CrossRef]
- Kooijmans, S.A.A.; Stremersch, S.; Braeckmans, K.; de Smedt, S.C.; Hendrix, A.; Wood, M.J.A.; Schiffelers, R.M.; Raemdonck, K.; Vader, P. Electroporation-induced siRNA precipitation obscures the efficiency of siRNA loading into extracellular vesicles. J. Control. Release 2013, 172, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Willis, G.R.; Kourembanas, S.; Mitsialis, S.A. Toward Exosome-Based Therapeutics: Isolation, Heterogeneity, and Fit-for-Purpose Potency. Front. Cardiovasc. Med. 2017, 4, 63. [Google Scholar] [CrossRef] [Green Version]
- Gangadaran, P.; Hong, C.M.; Oh, J.M.; Rajendran, R.L.; Kalimuthu, S.; Son, S.H.; Gopal, A.; Zhu, L.; Baek, S.H.; Jeong, S.Y.; et al. In vivo Non-invasive Imaging of Radio-Labeled Exosome-Mimetics Derived from Red Blood Cells in Mice. Front. Pharmacol. 2018, 9, 817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Son, S.H.; Oh, J.M.; Gangadaran, P.; Ji, H.D.; Lee, H.W.; Rajendran, R.L.; Baek, S.H.; Gopal, A.; Kalimuthu, S.; Jeong, S.Y.; et al. White blood cell labeling with Technetium-99m ((99m)Tc) using red blood cell extracellular vesicles-mimetics. Blood Cells Mol. Dis. 2020, 80, 102375. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.Y.; Ji, A.L.; Wang, Z.X.; Qiang, G.H.; Qu, Z.; Wu, J.H.; Jiang, C.P. Exosome-Mimetic Nanovesicles from Hepatocytes promote hepatocyte proliferation in vitro and liver regeneration in vivo. Sci. Rep. 2018, 8, 2471. [Google Scholar] [CrossRef] [Green Version]
- Kalimuthu, S.; Gangadaran, P.; Rajendran, R.L.; Zhu, L.; Oh, J.M.; Lee, H.W.; Gopal, A.; Baek, S.H.; Jeong, S.Y.; Lee, S.W.; et al. A New Approach for Loading Anticancer Drugs Into Mesenchymal Stem Cell-Derived Exosome Mimetics for Cancer Therapy. Front. Pharmacol. 2018, 9, 1116. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.T.; Umezaki, K.; Sawada, S.; Mukai, S.A.; Sasaki, Y.; Harada, N.; Shiku, H.; Akiyoshi, K. Engineering hybrid exosomes by membrane fusion with liposomes. Sci. Rep. 2016, 6, 21933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.; Wu, J.; Gu, W.; Huang, Y.; Tong, Z.; Huang, L.; Tan, J. Exosome-Liposome Hybrid Nanoparticles Deliver CRISPR/Cas9 System in MSCs. Adv. Sci. 2018, 5, 1700611. [Google Scholar] [CrossRef] [PubMed]
- Piffoux, M.; Silva, A.K.A.; Wilhelm, C.; Gazeau, F.; Tareste, D. Modification of Extracellular Vesicles by Fusion with Liposomes for the Design of Personalized Biogenic Drug Delivery Systems. ACS Nano 2018, 12, 6830–6842. [Google Scholar] [CrossRef]
- Rayamajhi, S.; Nguyen, T.D.T.; Marasini, R.; Aryal, S. Macrophage-derived exosome-mimetic hybrid vesicles for tumor targeted drug delivery. Acta Biomater. 2019, 94, 482–494. [Google Scholar] [CrossRef]
- Raimondo, S.; Naselli, F.; Fontana, S.; Monteleone, F.; Lo Dico, A.; Saieva, L.; Zito, G.; Flugy, A.; Manno, M.; Di Bella, M.A.; et al. Citrus limon-derived nanovesicles inhibit cancer cell proliferation and suppress CML xenograft growth by inducing TRAIL-mediated cell death. Oncotarget 2015, 6, 19514–19527. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, X.; Deng, Z.B.; Mu, J.; Zhang, L.; Yan, J.; Miller, D.; Feng, W.; McClain, C.J.; Zhang, H.G. Ginger-derived nanoparticles protect against alcohol-induced liver damage. J. Extracell. Vesicles 2015, 4, 28713. [Google Scholar] [CrossRef]
- Arntz, O.J.; Pieters, B.C.; Oliveira, M.C.; Broeren, M.G.; Bennink, M.B.; de Vries, M.; van Lent, P.L.; Koenders, M.I.; van den Berg, W.B.; van der Kraan, P.M.; et al. Oral administration of bovine milk derived extracellular vesicles attenuates arthritis in two mouse models. Mol. Nutr. Food Res. 2015, 59, 1701–1712. [Google Scholar] [CrossRef]
- Betker, J.L.; Angle, B.M.; Graner, M.W.; Anchordoquy, T.J. The Potential of Exosomes From Cow Milk for Oral Delivery. J. Pharm. Sci. 2019, 108, 1496–1505. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Chen, Y.; Wang, Q.; Jayasinghe, U.; Luo, X.; Wei, Q.; Wang, J.; Xiong, H.; Chen, C.; Xu, B.; et al. Exosome: Emerging biomarker in breast cancer. Oncotarget 2017, 8, 41717–41733. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Sun, X.; Chen, L. Exosome circ_0044516 promotes prostate cancer cell proliferation and metastasis as a potential biomarker. J. Cell Biochem. 2020, 121, 2118–2126. [Google Scholar] [CrossRef]
- Sharma, S.; Salomon, C. Techniques Associated with Exosome Isolation for Biomarker Development: Liquid Biopsies for Ovarian Cancer Detection. Methods Mol. Biol. 2020, 2055, 181–199. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Park, Y.H.; Jung, S.H.; Jang, S.H.; Kim, M.Y.; Lee, J.Y.; Chung, Y.J. Urinary exosome microRNA signatures as a noninvasive prognostic biomarker for prostate cancer. NPJ Genom. Med. 2021, 6, 45. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Hu, J.; Ye, Z.; Chen, S.; Chen, Y. Serum long noncoding RNA NNTAS1 protected by exosome is a potential biomarker and functions as an oncogene via the miR496/RAP2C axis in colorectal cancer. Mol. Med. Rep. 2021, 24, 585. [Google Scholar] [CrossRef]
- de la Torre Gomez, C.; Goreham, R.V.; Bech Serra, J.J.; Nann, T.; Kussmann, M. “Exosomics”-A Review of Biophysics, Biology and Biochemistry of Exosomes with a Focus on Human Breast Milk. Front. Genet. 2018, 9, 92. [Google Scholar] [CrossRef] [Green Version]
- Gezsi, A.; Kovacs, A.; Visnovitz, T.; Buzas, E.I. Systems biology approaches to investigating the roles of extracellular vesicles in human diseases. Exp. Mol. Med. 2019, 51, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Wolfers, J.; Lozier, A.; Raposo, G.; Regnault, A.; Thery, C.; Masurier, C.; Flament, C.; Pouzieux, S.; Faure, F.; Tursz, T.; et al. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat. Med. 2001, 7, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Wahlund, C.J.E.; Gucluler, G.; Hiltbrunner, S.; Veerman, R.E.; Naslund, T.I.; Gabrielsson, S. Exosomes from antigen-pulsed dendritic cells induce stronger antigen-specific immune responses than microvesicles in vivo. Sci. Rep. 2017, 7, 17095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luketic, L.; Delanghe, J.; Sobol, P.T.; Yang, P.; Frotten, E.; Mossman, K.L.; Gauldie, J.; Bramson, J.; Wan, Y. Antigen presentation by exosomes released from peptide-pulsed dendritic cells is not suppressed by the presence of active CTL. J. Immunol. 2007, 179, 5024–5032. [Google Scholar] [CrossRef]
- Kitai, Y.; Kawasaki, T.; Sueyoshi, T.; Kobiyama, K.; Ishii, K.J.; Zou, J.; Akira, S.; Matsuda, T.; Kawai, T. DNA-Containing Exosomes Derived from Cancer Cells Treated with Topotecan Activate a STING-Dependent Pathway and Reinforce Antitumor Immunity. J. Immunol. 2017, 198, 1649–1659. [Google Scholar] [CrossRef] [PubMed]
- Torralba, D.; Baixauli, F.; Villarroya-Beltri, C.; Fernandez-Delgado, I.; Latorre-Pellicer, A.; Acin-Perez, R.; Martin-Cofreces, N.B.; Jaso-Tamame, A.L.; Iborra, S.; Jorge, I.; et al. Priming of dendritic cells by DNA-containing extracellular vesicles from activated T cells through antigen-driven contacts. Nat. Commun. 2018, 9, 2658. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Types | Source | Features | Limitations | References |
|---|---|---|---|---|
| Cell-secreted exosomes | Human embryonic kidney (HEK) cells | Membrane resemblances to various tissues in our body Immunologically inert High efficiency in transfection | Low yield compared to body fluid and food-derived exosomes | [31,32] |
| Cancer cells | Tropism toward their parent cells | Less ideal pharmacokinetic profile Lack of studies for metastatic role of cancer cell exosomes | [33,34,35] | |
| Immune cells (e.g., dendritic cells) | Strictly defined molecular compositions Expression of NK cell-stimulating ligand peptides Surface modification using targeting moieties | Lack of understanding of DEX components (e.g., mRNAs, miRs, and cytokines) and mechanisms regarding how these factors interact with acceptor cells | [36,37,38,39,40] | |
| Stem cells (e.g., mesenchymal stem cell) | Easy availability from ethically acceptable tissues Large capacity for ex vivo expansion | Similar to other types of exosomes (e.g., lack of PK database and need to improve production efficiency) | [41,42,43] | |
| Blood-derived exosomes | Blood (red blood cells) | Relatively high yield from a single blood unit Reduced unexpected mutations from cell culture No occurring horizontal gene transfer Non-toxic/immunogenic by matching blood types High transfection efficiency | Not determined | [43,44] |
| Food-derived exosomes | Milk-derived exosomes | Dosing through less-invasive oral cavity Functionalized by simple post-insertion | Variation in shape, size, and cargo contents of exosomes depending on the diet and condition of the source Less understanding of the endogenous biological cargo of milk exosomes | [45,46,47,48] |
| Edible plants-derived exosomes (e.g., ginger, grapes, lemon, etc.) | Limited knowledge of cellular molecular properties of PDEs | [49,50,51] |
| Targeting Type | Examples | Target | Effect | Exosome Source | Delivery Molecule | Reference |
|---|---|---|---|---|---|---|
| Passive | CD47 surface decoration | SIRPα | Increased exosome circulation time | Mouse embryonic fibroblasts (MEFs) | mRNA | [130] |
| Surface PEGylation | N/A | Reduced exosome clearance Enhanced tumor penetration | 4T1 murine breast cancer cells | N/A | [131] | |
| iRGD peptide fusion with Lamp2b | N/A | Inhibited tumor growth without overt toxicity | Mouse immature dendritic cells | Doxorubicin | [132] | |
| Active | IL3-Lamp2b expressing exosome production | IL3 receptors | Increased intratumoral accumulation | HEK293T cells | Imatinib, BCR-ABL siRNA | [133] |
| Exosome azide integration, DBCO-PEG4-biotin avidin conjugation | Biotin receptors, glycan biosynthesis process | Higher uptake levels | B16F10 cells | Streptavidin-HRP | [134] | |
| Neuraminidase | Terminal sialic acid residues from glycoproteins | Rapid accumulation of EVs in the liver | MLP29 cells | N/A | [135] | |
| 3-(diethylamino) propyl-amine (DEAP) | N/A | pH sensitive uptake | RAW 264.7 cells | N/A | [136] | |
| αCD3 and αHER2 antibodies’ surface expression | CD3, HER2 receptors | Increased T cell activation | Expi293 cells | N/A | [137] | |
| anti-SSTR2 mAb | Somatostatin receptor 2 (SSTR2) | High toxicity of cancer cells | HEK 293 cells | Romidepsin | [138] | |
| DSPE-PEG biotin and avidin conjugation | Biotin receptors | Increased stability and encapsulation efficiency | HUVECs | N/A | [139] | |
| Diacyllipid–aptamer(sgc8) PEG conjugation | PTK7 | Increased cellular uptake | ImDC cells | Doxorubicin | [140] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.; Jang, H.; Cho, H.; Choi, J.; Hwang, K.Y.; Choi, Y.; Kim, S.H.; Yang, Y. Recent Advances in Exosome-Based Drug Delivery for Cancer Therapy. Cancers 2021, 13, 4435. https://doi.org/10.3390/cancers13174435
Kim H, Jang H, Cho H, Choi J, Hwang KY, Choi Y, Kim SH, Yang Y. Recent Advances in Exosome-Based Drug Delivery for Cancer Therapy. Cancers. 2021; 13(17):4435. https://doi.org/10.3390/cancers13174435
Chicago/Turabian StyleKim, Hyosuk, Hochung Jang, Haeun Cho, Jiwon Choi, Kwang Yeon Hwang, Yeonho Choi, Sun Hwa Kim, and Yoosoo Yang. 2021. "Recent Advances in Exosome-Based Drug Delivery for Cancer Therapy" Cancers 13, no. 17: 4435. https://doi.org/10.3390/cancers13174435
APA StyleKim, H., Jang, H., Cho, H., Choi, J., Hwang, K. Y., Choi, Y., Kim, S. H., & Yang, Y. (2021). Recent Advances in Exosome-Based Drug Delivery for Cancer Therapy. Cancers, 13(17), 4435. https://doi.org/10.3390/cancers13174435







