ncRNAs Orchestrate Chemosensitivity Induction by Neddylation Blockades
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. RNA Sequencing Meta-Analysis
2.2. Protein–Protein Interaction Prediction
2.3. Differential Expression Analysis and Heatmaps
2.4. Kaplan–Meier (KM) Survival Curves
2.5. Statistical Analysis
3. Results
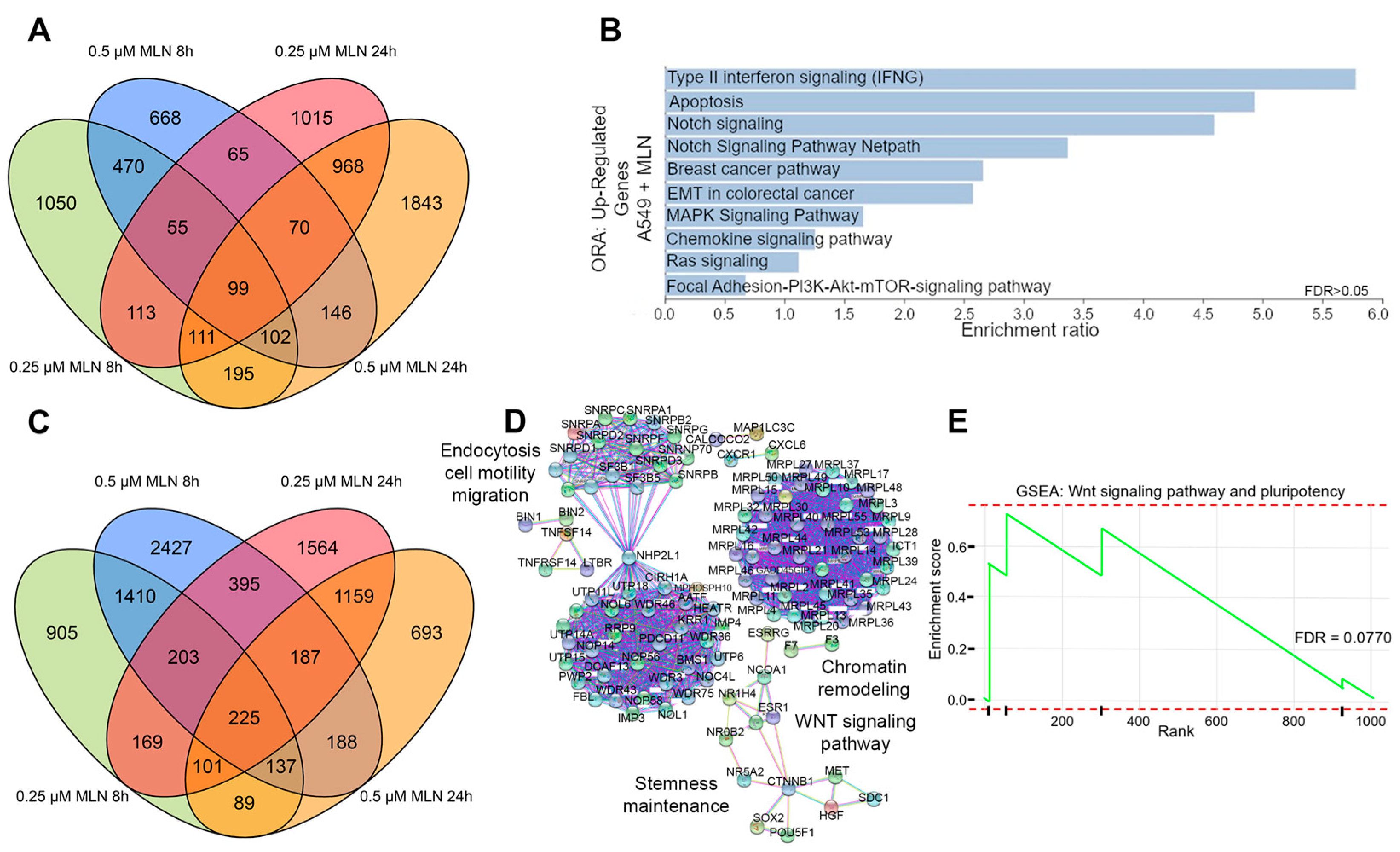
3.1. Neddylation Inhibition Reduces Stem Cell Maintenance and Survival in Lung Cancer Cells
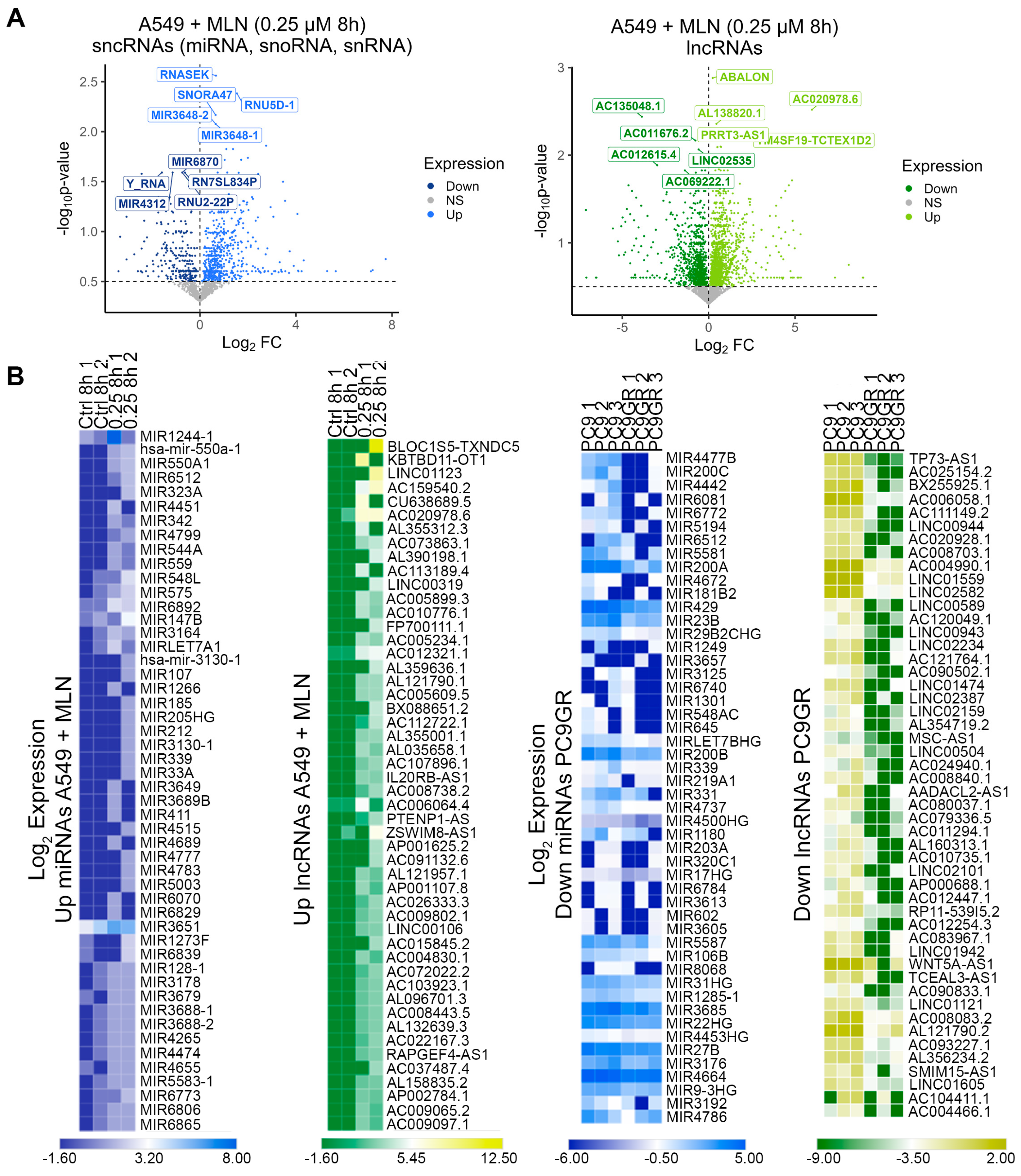
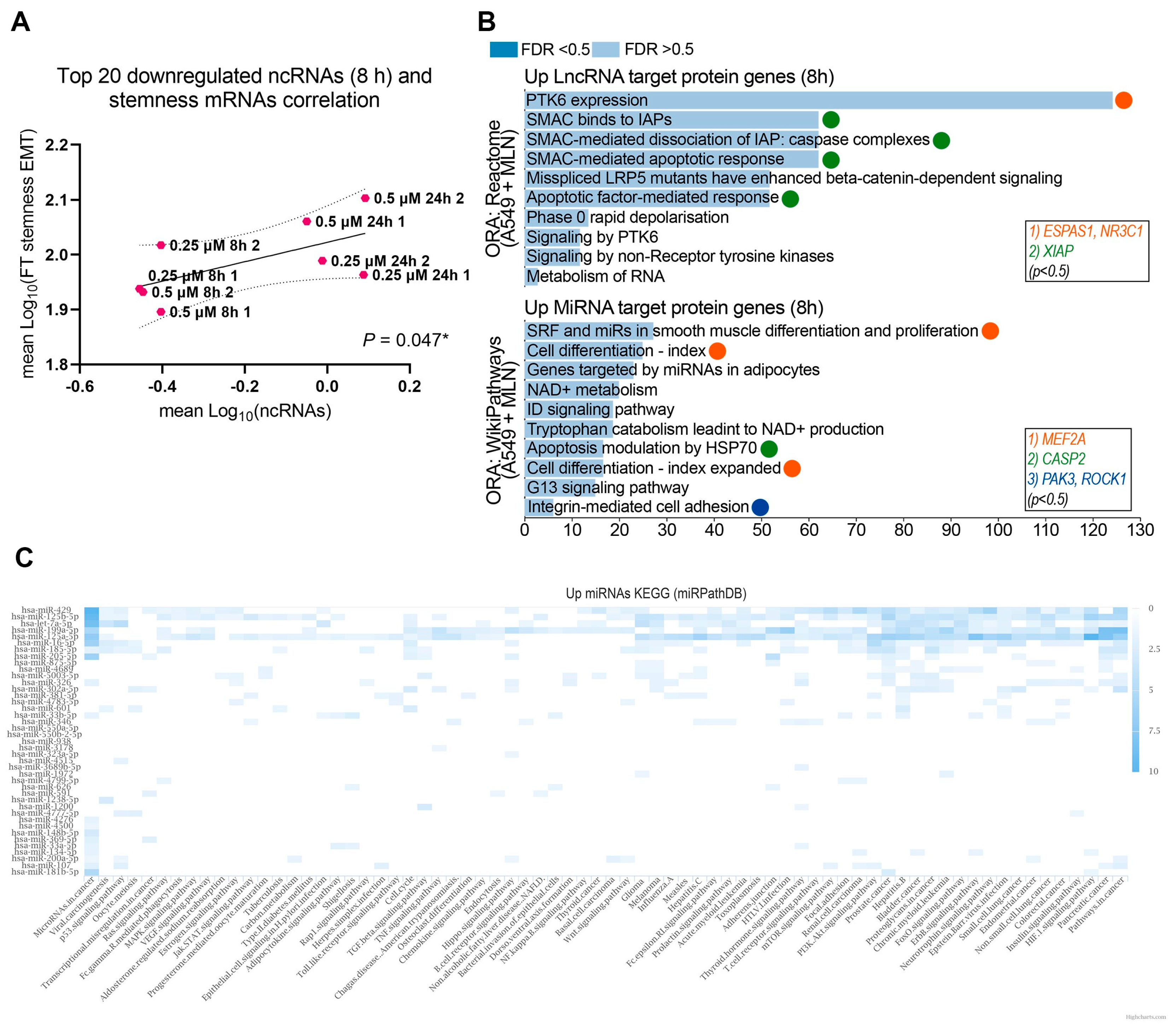
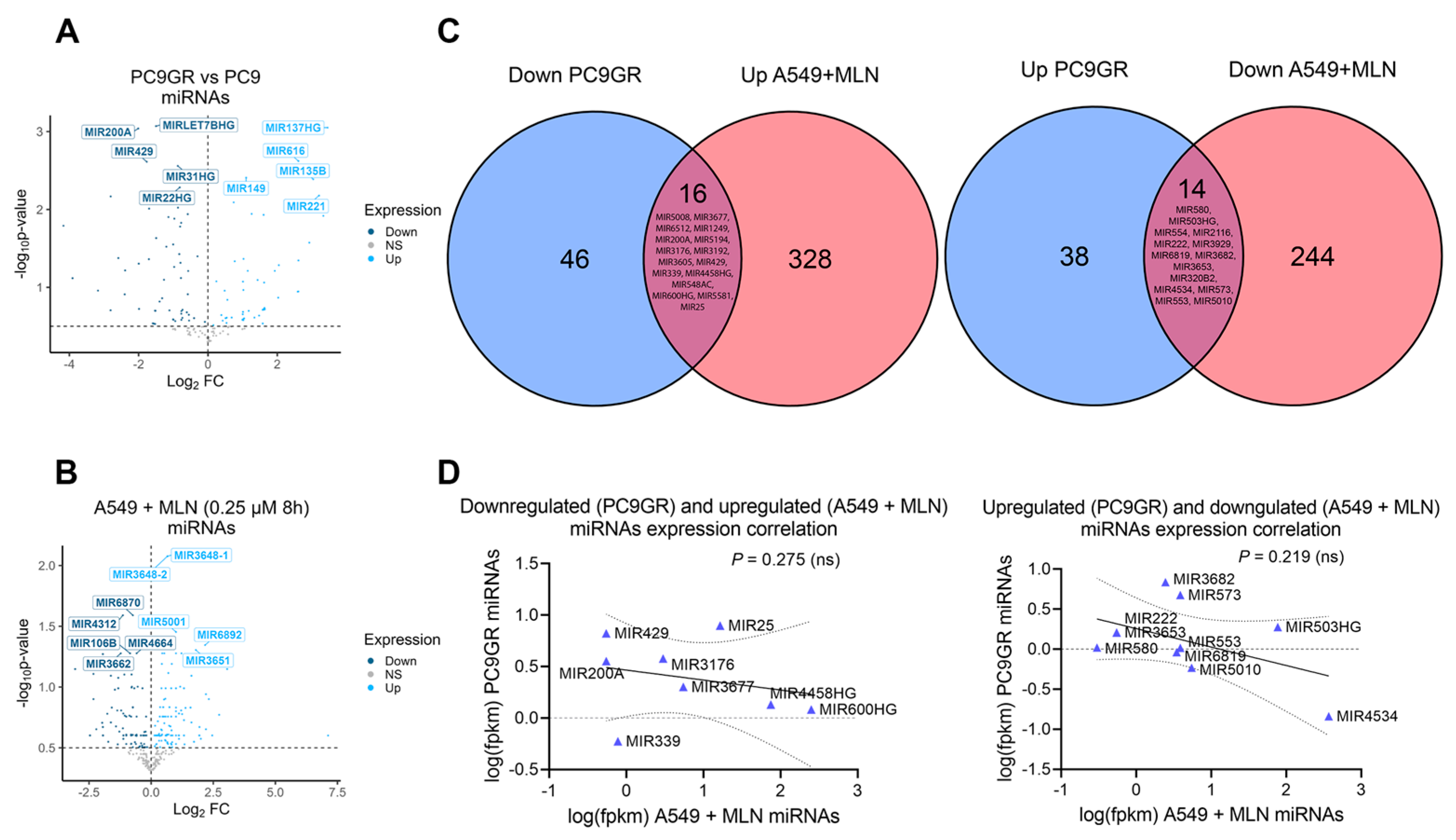
3.2. Reduced Cellular Stemness Correlates with ncRNA Differential Expression upon Neddylation Inhibition
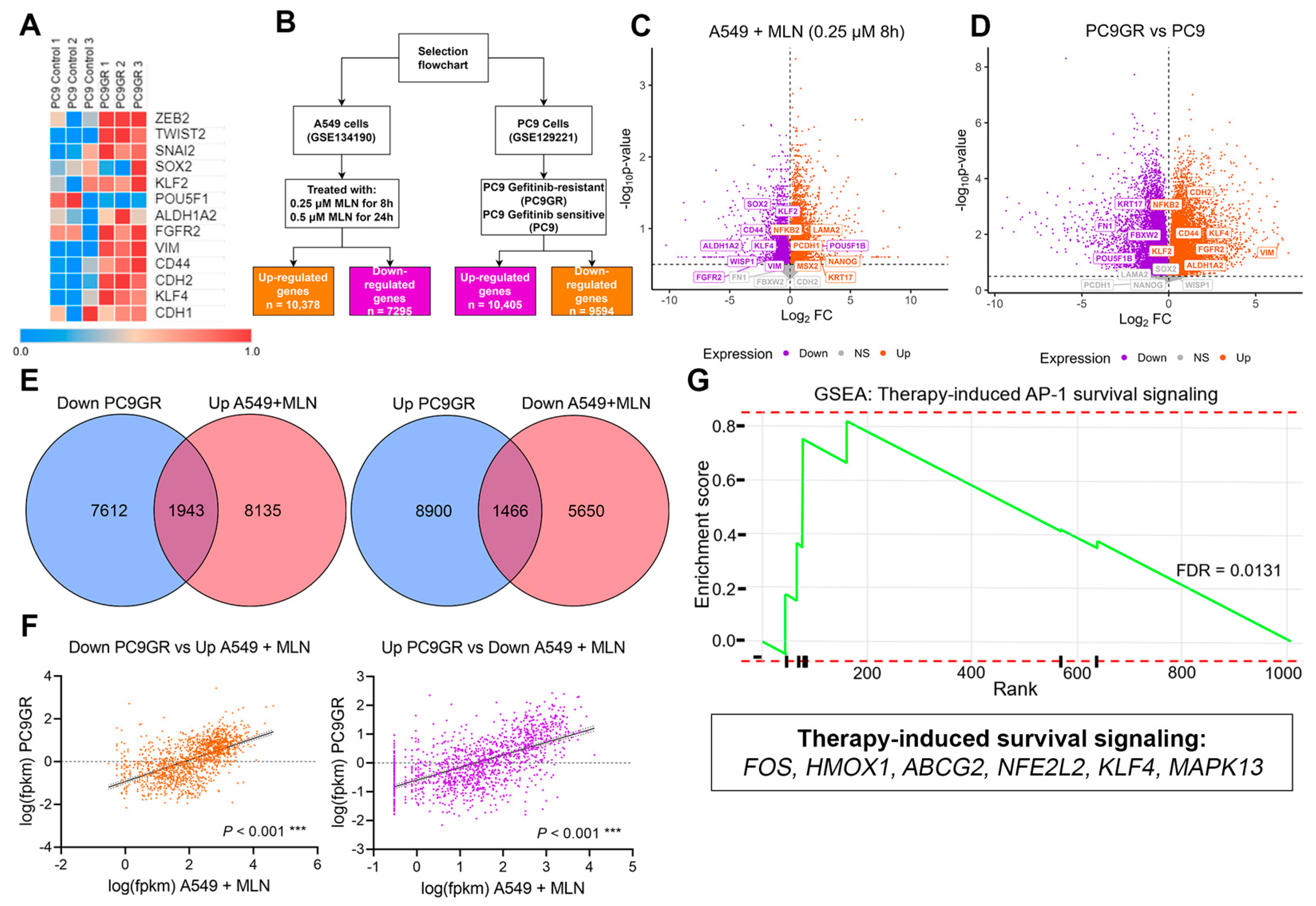
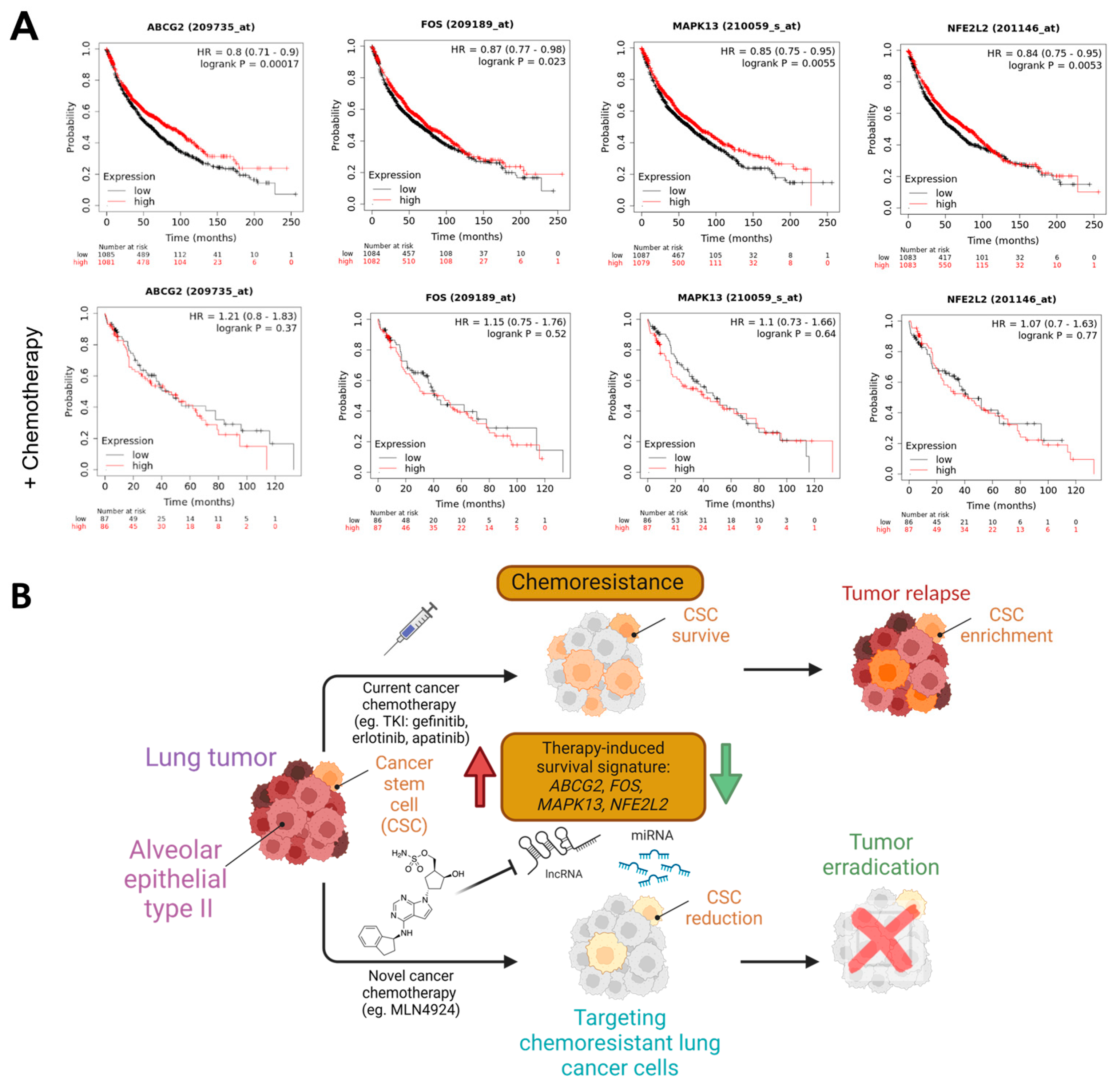
3.3. A Neddylation Inhibitor Attenuates Therapy-Induced Survival Activated by Chemoresistance in Lung Cancer Cells
3.4. Stemness and Epithelial–Mesenchymal Transition Factors Act as Neddylation Targets to Attenuate Chemoresistance Responses in Lung Cancer
4. Discussion and Perspectives
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Allemani, C.; Matsuda, T.; Di Carlo, V.; Harewood, R.; Matz, M.; Nikšić, M.; Bonaventure, A.; Valkov, M.; Johnson, C.J.; Estève, J.; et al. Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): Analysis of individual records for 37,513,025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 2018, 391, 1023–1075. [Google Scholar] [CrossRef] [PubMed]
- National Lung Screening Trial Research Team. The national lung screening trial: Overview and study design. Radiology 2011, 258, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Ning, J.; Ge, T.; Jiang, M.; Jia, K.; Wang, L.; Li, W.; Chen, B.; Liu, Y.; Wang, H.; Zhao, S.; et al. Early Diagnosis of Lung Cancer: Which Is the Optimal Choice? Aging 2021, 13, 6214. [Google Scholar] [CrossRef] [PubMed]
- Rubio, K.; Müller, J.M.; Mehta, A.; Watermann, I.; Olchers, T.; Koch, I.; Wessels, S.; Schneider, M.A.; Araujo-Ramos, T.; Singh, I.; et al. Preliminary results from the EMoLung clinical study showing early lung cancer detection by the LC score. Discov. Oncol. 2023, 14, 181. [Google Scholar] [CrossRef] [PubMed]
- Thai, A.A.; Solomon, B.J.; Sequist, L.V.; Gainor, J.F.; Heist, R.S. Lung cancer. Lancet 2021, 398, 535–554. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, R.M.; Sanchez, R. Lung cancer screening. Med. Clin. 2017, 101, 769–785. [Google Scholar] [CrossRef] [PubMed]
- Babar, L.; Modi, P.; Anjum, F. Lung Cancer Screening; Statpearls Publishing: St. Petersburg, FL, USA, 2019. [Google Scholar]
- Yang, C.-J.; Hwang, J.-J.; Kang, W.-Y.; Chong, I.-W.; Wang, T.-H.; Sheu, C.-C.; Tsai, J.-R.; Huang, M.-S. Gastro-intestinal metastasis of primary lung carcinoma: Clinical presentations and outcome. Lung Cancer 2006, 54, 319–323. [Google Scholar] [CrossRef]
- Sakai, H.; Egi, H.; Hinoi, T.; Tokunaga, M.; Kawaguchi, Y.; Shinomura, M.; Adachi, T.; Arihiro, K.; Ohdan, H. Primary lung cancer presenting with metastasis to the colon: A case report. World J. Surg. Oncol. 2012, 10, 1–5. [Google Scholar] [CrossRef]
- Vallières, E. Early stage small cell lung cancer: Very rare, but a surgical disease! J. Thorac. Dis. 2017, 9, 4185. [Google Scholar] [CrossRef] [PubMed]
- Krpina, K.; Vranić, S.; Tomić, K.; Samaržija, M.; Batičić, L. Small Cell Lung Carcinoma: Current Diagnosis, Biomarkers, and Treatment Options with Future Perspectives. Biomedicines 2023, 11, 1982. [Google Scholar] [CrossRef]
- Rubio, K.; Romero-Olmedo, A.J.; Sarvari, P.; Swaminathan, G.; Ranvir, V.P.; Rogel-Ayala, D.G.; Cordero, J.; Günther, S.; Mehta, A.; Bassaly, B.; et al. Non-canonical integrin signaling activates EGFR and RAS-MAPK-ERK signaling in small cell lung cancer. Theranostics 2023, 13, 2384. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, K.D.; Berns, A. Cell of origin of lung cancer. Mol. Oncol. 2010, 4, 397–403. [Google Scholar] [CrossRef]
- Herbst, R.S.; Heymach, J.V.; Lippman, S.M. Lung cancer. N. Engl. J. Med. 2008, 359, 1367–1380. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-B.; Dunn, C.T.; Park, K.-S. Recent progress in mapping the emerging landscape of the small-cell lung cancer genome. Exp. Mol. Med. 2019, 51, 1–13. [Google Scholar] [CrossRef]
- Marshall, E.A.; Ng, K.W.; Kung, S.H.; Conway, E.M.; Martinez, V.D.; Halvorsen, E.C.; Rowbotham, D.A.; Vucic, E.A.; Plumb, A.W.; Becker-Santos, D.D. Emerging roles of T helper 17 and regulatory T cells in lung cancer progression and metastasis. Mol. Cancer 2016, 15, 1–15. [Google Scholar] [CrossRef]
- Blandin Knight, S.; Crosbie, P.A.; Balata, H.; Chudziak, J.; Hussell, T.; Dive, C. Progress and prospects of early detection in lung cancer. Open Biol. 2017, 7, 170070. [Google Scholar] [CrossRef]
- Zappa, C.; Mousa, S.A. Non-small cell lung cancer: Current treatment and future advances. Transl. Lung Cancer Res. 2016, 5, 288. [Google Scholar] [CrossRef]
- Myers, D.J.; Wallen, J.M. Lung Adenocarcinoma; StatPearls Publishing: St. Petersburg, FL, USA, 2023. [Google Scholar]
- Lee, M.C.; Cai, H.; Murray, C.W.; Li, C.; Shue, Y.T.; Andrejka, L.; He, A.L.; Holzem, A.M.; Drainas, A.P.; Ko, J.H. A multiplexed in vivo approach to identify driver genes in small cell lung cancer. Cell Rep. 2023, 42, 111990. [Google Scholar] [CrossRef]
- Ju, F.; Atyah, M.M.; Horstmann, N.; Gul, S.; Vago, R.; Bruns, C.J.; Zhao, Y.; Dong, Q.-Z.; Ren, N. Characteristics of the cancer stem cell niche and therapeutic strategies. Stem Cell Res. Ther. 2022, 13, 1–17. [Google Scholar] [CrossRef]
- Yang, L.; Shi, P.; Zhao, G.; Xu, J.; Peng, W.; Zhang, J.; Zhang, G.; Wang, X.; Dong, Z.; Chen, F. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target. Ther. 2020, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Feng, J.; Zhang, Q.; He, Y.; Xu, C.; Wang, C.; Li, W. Epigenetic regulation of stem cells in lung cancer oncogenesis and therapy resistance. Front. Genet. 2023, 14, 1120815. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, W.; Sun, Y.; Jia, L. Protein neddylation and its alterations in human cancers for targeted therapy. Cell. Signal. 2018, 44, 92–102. [Google Scholar] [CrossRef]
- Li, L.; Kang, J.; Zhang, W.; Cai, L.; Wang, S.; Liang, Y.; Jiang, Y.; Liu, X.; Zhang, Y.; Ruan, H.; et al. Validation of NEDD8-conjugating enzyme UBC12 as a new therapeutic target in lung cancer. EBioMedicine 2019, 45, 81–91. [Google Scholar] [CrossRef]
- Wang, S.; Xian, J.; Li, L.; Jiang, Y.; Liu, Y.; Cai, L.; Hoffman, R.M.; Jia, L.; Zhao, H.; Zhang, Y.; et al. NEDD8-conjugating enzyme UBC12 as a novel therapeutic target in esophageal squamous cell carcinoma. Signal Transduct. Target. Ther. 2020, 5, 123. [Google Scholar] [CrossRef]
- Zhou, W.; Xu, J.; Li, H.; Xu, M.; Chen, Z.J.; Wei, W.; Pan, Z.; Sun, Y. Neddylation E2 UBE2F promotes the survival of lung cancer cells by activating CRL5 to degrade NOXA via the K11 linkage. Clin. Cancer Res. 2017, 23, 1104–1116. [Google Scholar] [CrossRef]
- Zhao, Y.; Morgan, M.A.; Sun, Y.J.A. Targeting Neddylation pathways to inactivate cullin-RING ligases for anticancer therapy. Antioxid. Redox Signal. 2014, 21, 2383–2400. [Google Scholar] [CrossRef]
- Xu, T.; Ma, Q.; Li, Y.; Yu, Q.; Pan, P.; Zheng, Y.; Li, Z.; Xiong, X.; Hou, T.; Yu, B.; et al. A small molecule inhibitor of the UBE2F-CRL5 axis induces apoptosis and radiosensitization in lung cancer. Signal Transduct. Target. Ther. 2022, 7, 354. [Google Scholar] [CrossRef] [PubMed]
- Govindarajulu, M.; Ramesh, S.; Shankar, T.; Kora, M.K.; Moore, T.; Dhanasekaran, M. Role of Neddylation in Neurodegenerative Diseases. NeuroSci 2022, 3, 533–545. [Google Scholar] [CrossRef]
- Yao, J.; Liang, X.; Liu, Y.; Zheng, M. Neddylation: A versatile pathway takes on chronic liver diseases. Front. Med. 2020, 7, 586881. [Google Scholar] [CrossRef]
- Kamitani, T.; Kito, K.; Nguyen, H.P.; Yeh, E.T. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J. Biol. Chemisrty 1997, 272, 28557–28562. [Google Scholar] [CrossRef]
- Milhollen, M.A.; Traore, T.; Adams-Duffy, J.; Thomas, M.P.; Berger, A.J.; Dang, L.; Dick, L.R.; Garnsey, J.J.; Koenig, E.; Langston, S.P. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: Rationale for treatment of NF-κB–dependent lymphoma. J. Am. Soc. Hematol. 2010, 116, 1515–1523. [Google Scholar] [CrossRef]
- Kumar, D.; Das, M.; Sauceda, C.; Ellies, L.G.; Kuo, K.; Parwal, P.; Kaur, M.; Jih, L.; Bandyopadhyay, G.K.; Burton, D.; et al. Degradation of splicing factor SRSF3 contributes to progressive liver disease. J. Clin. Investig. 2019, 129, 4477–4491. [Google Scholar] [CrossRef]
- Zubiete-Franco, I.; Fernández-Tussy, P.; Barbier-Torres, L.; Simon, J.; Fernández-Ramos, D.; Lopitz-Otsoa, F.; Gutiérrez-de Juan, V.; de Davalillo, S.L.; Duce, A.M.; Iruzubieta, P.; et al. Deregulated neddylation in liver fibrosis. Hepatology 2017, 65, 694–709. [Google Scholar] [CrossRef] [PubMed]
- Mathewson, N.; Toubai, T.; Kapeles, S.; Sun, Y.; Oravecz-Wilson, K.; Tamaki, H.; Wang, Y.; Hou, G.; Sun, Y.; Reddy, P. Neddylation plays an important role in the regulation of murine and human dendritic cell function. J. Am. Soc. Hematol. 2013, 122, 2062–2073. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Shang, Z.; Li, S.; Gao, P.; Zhang, Y.; Hou, S.; Qin, P.; Dong, Z.; Hu, T.; Chen, P. Neddylation inhibitor MLN4924 induces G2 cell cycle arrest, DNA damage and sensitizes esophageal squamous cell carcinoma cells to cisplatin. Oncol. Lett. 2018, 15, 2583–2589. [Google Scholar] [CrossRef] [PubMed]
- Soucy, T.A.; Smith, P.G.; Milhollen, M.A.; Berger, A.J.; Gavin, J.M.; Adhikari, S.; Brownell, J.E.; Burke, K.E.; Cardin, D.P.; Critchley, S. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 2009, 458, 732–736. [Google Scholar] [CrossRef]
- Brownell, J.E.; Sintchak, M.D.; Gavin, J.M.; Liao, H.; Bruzzese, F.J.; Bump, N.J.; Soucy, T.A.; Milhollen, M.A.; Yang, X.; Burkhardt, A.L.; et al. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: The NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol. Cell 2010, 37, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Kuo, K.-L.; Ho, I.-L.; Shi, C.-S.; Wu, J.-T.; Lin, W.-C.; Tsai, Y.-C.; Chang, H.-C.; Chou, C.-T.; Hsu, C.-H.; Hsieh, J.-T.; et al. MLN4924, a novel protein neddylation inhibitor, suppresses proliferation and migration of human urothelial carcinoma: In vitro and in vivo studies. Cancer Lett. 2015, 363, 127–136. [Google Scholar] [CrossRef]
- Lan, H.; Tang, Z.; Jin, H.; Sun, Y. Neddylation inhibitor MLN4924 suppresses growth and migration of human gastric cancer cells. Sci. Rep. 2016, 6, 24218. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.; Si, Y.; Yu, H.; Zhang, L.; Xie, P.; Jiang, W. MLN4924 (Pevonedistat), a protein neddylation inhibitor, suppresses proliferation and migration of human clear cell renal cell carcinoma. Sci. Rep. 2017, 7, 5599. [Google Scholar] [CrossRef]
- Wang, Y.; Luo, Z.; Pan, Y.; Wang, W.; Zhou, X.; Jeong, L.S.; Chu, Y.; Liu, J.; Jia, L. Targeting protein neddylation with an NEDD8-activating enzyme inhibitor MLN4924 induced apoptosis or senescence in human lymphoma cells. Cancer Biol. Ther. 2015, 16, 420–429. [Google Scholar] [CrossRef]
- Han, K.; Wang, Q.; Cao, H.; Qiu, G.; Cao, J.; Li, X.; Wang, J.; Shen, B.; Zhang, J. The NEDD8-activating enzyme inhibitor MLN4924 induces G2 arrest and apoptosis in T-cell acute lymphoblastic leukemia. Oncotarget 2016, 7, 23812. [Google Scholar] [CrossRef]
- Bahjat, M.; de Wilde, G.; van Dam, T.; Maas, C.; Bloedjes, T.; Bende, R.J.; van Noesel, C.J.; Luijks, D.M.; Eldering, E.; Kersten, M.J. The NEDD8-activating enzyme inhibitor MLN4924 induces DNA damage in Ph+ leukemia and sensitizes for ABL kinase inhibitors. Cell Cycle 2019, 18, 2307–2322. [Google Scholar] [CrossRef]
- Chen, Y.; Sun, L. Inhibition of NEDD8 NEDDylation induced apoptosis in acute myeloid leukemia cells via p53 signaling pathway. Biosci. Rep. 2022, 42, BSR20220994. [Google Scholar] [CrossRef]
- Milhollen, M.; Narayanan, U.; Duffy, J.; Amidon, B.; Soucy, T.A.; Berger, A.J.; Langston, S.P.; Sells, T.; Smith, P.G. MLN4924, a potent and novel small molecule inhibitor of Nedd8 activating enzyme, induces DNA re-replication and apoptosis in cultured human tumor cells. Blood 2008, 112, 3621. [Google Scholar] [CrossRef]
- Lv, Y.; Li, B.; Han, K.; Xiao, Y.; Yu, X.; Ma, Y.; Jiao, Z.; Gao, J. The Nedd8-activating enzyme inhibitor MLN4924 suppresses colon cancer cell growth via triggering autophagy. Korean J. Physiol. Pharmacol. 2018, 22, 617–625. [Google Scholar] [CrossRef]
- Rellinger, E.J.; Padmanabhan, C.; Qiao, J.; Appert, A.; Waterson, A.G.; Lindsley, C.W.; Beauchamp, R.D.; Chung, D.H. ML327 induces apoptosis and sensitizes Ewing sarcoma cells to TNF-related apoptosis-inducing ligand. Biochem. Biophys. Res. Commun. 2017, 491, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.-S.; Kuo, K.-L.; Lin, W.-C.; Chen, M.-S.; Liu, S.-H.; Liao, S.-M.; Hsu, C.-H.; Chang, Y.-W.; Chang, H.-C.; Huang, K.-H. Neddylation inhibitor, MLN4924 suppresses angiogenesis in huvecs and solid cancers: In vitro and in vivo study. Am. J. Cancer Res. 2020, 10, 953. [Google Scholar] [PubMed]
- Bhatia, S.; Pavlick, A.C.; Boasberg, P.; Thompson, J.A.; Mulligan, G.; Pickard, M.D.; Faessel, H.; Dezube, B.J.; Hamid, O. A phase I study of the investigational NEDD8-activating enzyme inhibitor pevonedistat (TAK-924/MLN4924) in patients with metastatic melanoma. Investig. New Drugs 2016, 34, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Shah, J.J.; Jakubowiak, A.J.; O’Connor, O.A.; Orlowski, R.Z.; Harvey, R.D.; Smith, M.R.; Lebovic, D.; Diefenbach, C.; Kelly, K.; Hua, Z.; et al. Phase I study of the novel investigational NEDD8-activating enzyme inhibitor pevonedistat (MLN4924) in patients with relapsed/refractory multiple myeloma or lymphoma. Clin. Cancer Res. 2016, 22, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Sarantopoulos, J.; Shapiro, G.I.; Cohen, R.B.; Clark, J.W.; Kauh, J.S.; Weiss, G.J.; Cleary, J.M.; Mahalingam, D.; Pickard, M.D.; Faessel, H.M.; et al. Phase I study of the investigational NEDD8-activating enzyme inhibitor pevonedistat (TAK-924/MLN4924) in patients with advanced solid tumors. Clin. Cancer Res. 2016, 22, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Nomura, Y.; Nakano, M.; Woo Sung, H.; Han, M.; Pandey, D. Inhibition of HDAC6 activity protects against endothelial dysfunction and atherogenesis in vivo: A role for HDAC6 neddylation. Front. Physiol. 2021, 12, 675724. [Google Scholar] [CrossRef]
- Song, Y.-A.; Ma, T.; Zhang, X.-Y.; Cheng, X.-S.; Olajuyin, A.-M.; Sun, Z.-F.; Zhang, X.-J. Apatinib preferentially inhibits PC9 gefitinib-resistant cancer cells by inducing cell cycle arrest and inhibiting VEGFR signaling pathway. Cancer Cell Int. 2019, 19, 117. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.-F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, M.; Yu, G.; Chen, P.; Li, H.; Wei, D.; Zhu, J.; Xie, L.; Jia, H.; Shi, J.; et al. Overactivated neddylation pathway as a therapeutic target in lung cancer. J. Natl. Cancer Inst. 2014, 106, dju083. [Google Scholar] [CrossRef]
- Wei, N.; Song, Y.a.; Zhang, F.; Sun, Z.; Zhang, X. Transcriptome profiling of acquired gefitinib resistant lung cancer cells reveals dramatically changed transcription programs and new treatment targets. Front. Oncol. 2020, 10, 1424. [Google Scholar] [CrossRef]
- Győrffy, B.; Surowiak, P.; Budczies, J.; Lánczky, A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE 2013, 8, e82241. [Google Scholar] [CrossRef]
- Xirodimas, D.P. Novel substrates and functions for the ubiquitin-like molecule NEDD8. Transactions 2008, 36, 802–806. [Google Scholar] [CrossRef]
- Gan-Erdene, T.; Nagamalleswari, K.; Yin, L.; Wu, K.; Pan, Z.-Q.; Wilkinson, K.D. Identification and characterization of DEN1, a deneddylase of the ULP family. J. Biol. Chem. 2003, 278, 28892–28900. [Google Scholar] [CrossRef]
- Kandala, S.; Kim, I.M.; Su, H. Neddylation and deneddylation in cardiac biology. Am. J. Cardiovasc. Dis. 2014, 4, 140. [Google Scholar]
- Gong, L.; Yeh, E.T. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J. Biol. Chem. 1999, 274, 12036–12042. [Google Scholar] [CrossRef]
- Pan, Z.-Q.; Kentsis, A.; Dias, D.C.; Yamoah, K.; Wu, K. Nedd8 on cullin: Building an expressway to protein destruction. Oncogene 2004, 23, 1985–1997. [Google Scholar] [CrossRef]
- Saha, A.; Deshaies, R. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol. Cell 2008, 32, 21–31. [Google Scholar] [CrossRef]
- Abbas, T.; Sivaprasad, U.; Terai, K.; Amador, V.; Pagano, M.; Dutta, A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008, 22, 2496–2506. [Google Scholar] [CrossRef]
- Rada, P.; Rojo, A.I.; Chowdhry, S.; McMahon, M.; Hayes, J.D.; Cuadrado, A. SCF/β-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol. Cell. Biol. 2011, 31, 1121–1133. [Google Scholar] [CrossRef] [PubMed]
- Rada, P.; Rojo, A.I.; Evrard-Todeschi, N.; Innamorato, N.G.; Cotte, A.; Jaworski, T.; Tobón-Velasco, J.C.; Devijver, H.; García-Mayoral, M.F.; Van Leuven, F.; et al. Structural and functional characterization of Nrf2 degradation by the glycogen synthase kinase 3/β-TrCP axis. Mol. Cell. Biol. 2012, 32, 3486–3499. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; McCall, C.M.; Ohta, T.; Xiong, Y. Targeted ubiquitination of CDT1 by the DDB1–CUL4A–ROC1 ligase in response to DNA damage. Nat. Cell Biol. 2004, 6, 1003–1009. [Google Scholar] [CrossRef]
- Welcker, M.; Orian, A.; Jin, J.; Grim, J.A.; Harper, J.W.; Eisenman, R.N.; Clurman, B.E. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. USA 2004, 101, 9085–9090. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Li, Y.; Yang, R.; Xi, N.; Sun, Y. Inactivation of SAG E3 ubiquitin ligase blocks embryonic stem cell differentiation and sensitizes leukemia cells to retinoid acid. PLoS ONE 2011, 6, e27726. [Google Scholar] [CrossRef]
- Nawrocki, S.T.; Kelly, K.R.; Smith, P.G.; Espitia, C.M.; Possemato, A.; Beausoleil, S.A.; Milhollen, M.; Blakemore, S.; Thomas, M.; Berger, A.; et al. Disrupting protein NEDDylation with MLN4924 is a novel strategy to target cisplatin resistance in ovarian cancer. Clin. Cancer Res. 2013, 19, 3577–3590. [Google Scholar] [CrossRef]
- Paiva, C.; Godbersen, J.; Berger, A.; Brown, J.; Danilov, A.V. Targeting neddylation induces DNA damage and checkpoint activation and sensitizes chronic lymphocytic leukemia B cells to alkylating agents. Cell Death Dis. 2015, 6, e1807. [Google Scholar] [CrossRef]
- Ho, I.-L.; Kuo, K.-L.; Liu, S.-H.; Chang, H.-C.; Hsieh, J.-T.; Wu, J.-T.; Chiang, C.-K.; Lin, W.-C.; Tsai, Y.-C.; Chou, C.-T.; et al. MLN4924 synergistically enhances cisplatin-induced cytotoxicity via JNK and Bcl-xL pathways in human urothelial carcinoma. Sci. Rep. 2015, 5, 16948. [Google Scholar] [CrossRef]
- Lin, W.-C.; Kuo, K.-L.; Shi, C.-S.; Wu, J.-T.; Hsieh, J.-T.; Chang, H.-C.; Liao, S.-M.; Chou, C.-T.; Chiang, C.-K.; Chiu, W.-S.; et al. MLN4924, a Novel NEDD8-activating enzyme inhibitor, exhibits antitumor activity and enhances cisplatin-induced cytotoxicity in human cervical carcinoma: In vitro and in vivo study. Am. J. Cancer Res. 2015, 5, 3350. [Google Scholar] [PubMed]
- Zhou, L.; Chen, S.; Zhang, Y.; Kmieciak, M.; Leng, Y.; Li, L.; Lin, H.; Rizzo, K.A.; Dumur, C.I.; Ferreira-Gonzalez, A.; et al. The NAE inhibitor pevonedistat interacts with the HDAC inhibitor belinostat to target AML cells by disrupting the DDR. Blood J. Am. Soc. Hematol. 2016, 127, 2219–2230. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Liang, Y.; Li, L.; Wang, X.; Yan, Z.; Dong, C.; Zeng, M.S.; Zhong, Q.; Liu, X.K.; Yu, J.; et al. The Nedd8-activating enzyme inhibitor MLN 4924 (TAK-924/Pevonedistat) induces apoptosis via c-Myc-Noxa axis in head and neck squamous cell carcinoma. Cell Prolif. 2019, 52, e12536. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.-H.; Lee, C.-Y.; Huang, T.-J.; Huang, K.-Y.; Tang, C.-H.; Liu, S.-H.; Kuo, K.-L.; Kuan, F.-C.; Lin, W.-C.; Shi, C.-S. MLN4924, a protein neddylation inhibitor, suppresses the growth of human chondrosarcoma through inhibiting cell proliferation and inducing endoplasmic reticulum stress-related apoptosis. Int. J. Mol. Sci. 2018, 20, 72. [Google Scholar] [CrossRef] [PubMed]
- Ai, T.-J.; Sun, J.-Y.; Du, L.-J.; Shi, C.; Li, C.; Sun, X.-N.; Liu, Y.; Li, L.; Xia, Z.; Jia, L.; et al. Inhibition of neddylation by MLN4924 improves neointimal hyperplasia and promotes apoptosis of vascular smooth muscle cells through p53 and p62. Cell Death Differ. 2018, 25, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Jiang, Y.; Wu, J.; Zhang, W.; Liang, Y.; Jia, L.; Yu, J.; Jeong, L.; Li, L. NEDD8-activating enzyme inhibitor, MLN4924 (Pevonedistat) induces NOXA-dependent apoptosis through up-regulation of ATF-4. Biochem. Biophys. Res. Commun. 2017, 488, 1–5. [Google Scholar] [CrossRef]
- Knorr, K.; Schneider, P.; Meng, X.; Dai, H.; Smith, B.; Hess, A.; Karp, J.; Kaufmann, S.H. MLN4924 induces Noxa upregulation in acute myelogenous leukemia and synergizes with Bcl-2 inhibitors. Cell Death Differ. 2015, 22, 2133–2142. [Google Scholar] [CrossRef]
- Zhou, Q.; Li, H.; Li, Y.; Tan, M.; Fan, S.; Cao, C.; Meng, F.; Zhu, L.; Zhao, L.; Guan, M.-X.; et al. Inhibiting neddylation modification alters mitochondrial morphology and reprograms energy metabolism in cancer cells. JCI Insight 2019, 4, e121582. [Google Scholar] [CrossRef]
- Fu, D.-J.; Wang, T. Targeting NEDD8-activating enzyme for cancer therapy: Developments, clinical trials, challenges and future research directions. J. Hematol. Oncol. 2023, 16, 87. [Google Scholar] [CrossRef]
- Barbier-Torres, L.; Delgado, T.C.; García-Rodríguez, J.L.; Zubiete-Franco, I.; Fernandez-Ramos, D.; Buqué, X.; Cano, A.; Gutierrez-de Juan, V.; Fernández-Domínguez, I.; Lopitz-Otsoa, F.; et al. Stabilization of LKB1 and Akt by neddylation regulates energy metabolism in liver cancer. Oncotarget 2015, 6, 2509. [Google Scholar] [CrossRef]
- Zhang, Y.; Shi, C.-C.; Zhang, H.-P.; Li, G.-Q.; Li, S.-S. MLN4924 suppresses neddylation and induces cell cycle arrest, senescence, and apoptosis in human osteosarcoma. Oncotarget 2016, 7, 45263. [Google Scholar] [CrossRef]
- Zhou, X.; Tan, M.; Nyati, M.K.; Zhao, Y.; Wang, G.; Sun, Y. Blockage of neddylation modification stimulates tumor sphere formation in vitro and stem cell differentiation and wound healing in vivo. Proc. Natl. Acad. Sci. USA 2016, 113, E2935–E2944. [Google Scholar] [CrossRef]
- Park, S.Y.; Park, J.-W.; Lee, G.-W.; Li, L.; Chun, Y.-S. Inhibition of neddylation facilitates cell migration through enhanced phosphorylation of caveolin-1 in PC3 and U373MG cells. BMC Cancer 2018, 18, 1–11. [Google Scholar] [CrossRef]
- Onodera, K.; Sakurada, A.; Notsuda, H.; Watanabe, T.; Matsuda, Y.; Noda, M.; Endo, C.; Okada, Y. Growth inhibition of KRAS- and EGFR-mutant lung adenocarcinoma by cosuppression of STAT3 and the SRC/ARHGAP35 axis. Oncol. Rep. 2018, 40, 1761–1768. [Google Scholar] [CrossRef]
- Orabi, M.A.; Alqahtani, O.S.; Alyami, B.A.; Al Awadh, A.A.; Abdel-Sattar, E.S.; Matsunami, K.; Hamdan, D.I.; Abouelela, M.E. Human Lung Cancer (A549) Cell Line Cytotoxicity and Anti-Leishmania major Activity of Carissa macrocarpa Leaves: A Study Supported by UPLC-ESI-MS/MS Metabolites Profiling and Molecular Docking. Pharmaceuticals 2022, 15, 1561. [Google Scholar] [CrossRef] [PubMed]
- Park, M.Y.; Jung, M.H.; Eo, E.Y.; Kim, S.; Lee, S.H.; Lee, Y.J.; Park, J.S.; Cho, Y.J.; Chung, J.H.; Kim, C.H.; et al. Generation of lung cancer cell lines harboring EGFR T790M mutation by CRISPR/Cas9-mediated genome editing. Oncotarget 2017, 8, 36331–36338. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.S.H.; Kobayashi, S.; Costa, D.B. Acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancers dependent on the epidermal growth factor receptor pathway. Clin Lung Cancer 2009, 10, 281–289. [Google Scholar] [CrossRef]
- Pan, J.; Li, M.; Yu, F.; Zhu, F.; Wang, L.; Ning, D.; Hou, X.; Jiang, F. Up-Regulation of p53/miR-628-3p Pathway, a Novel Mechanism of Shikonin on Inhibiting Proliferation and Inducing Apoptosis of A549 and PC-9 Non-Small Cell Lung Cancer Cell Lines. Front. Pharmacol. 2021, 12, 766165. [Google Scholar] [CrossRef]
- Zhao, Y.M.; Su, B.; Yang, X.J.; Shi, J.Y.; Tang, L.; Zhang, J.; Li, J.Y.; Chen, J. Small molecule inhibitor SB203580 enhances the antitumor effect of gefitinib in PC-9 and A549 lung cancer cell lines. Zhonghua Zhong Liu Za Zhi [Chin. J. Oncol.] 2013, 35, 103–108. [Google Scholar] [CrossRef]
- El-Mesery, M.; Anany, M.A.; Hazem, S.H.; Shaker, M.E. The NEDD8-activating enzyme inhibition with MLN4924 sensitizes human cancer cells of different origins to apoptosis and necroptosis. Arch. Biochem. Biophys. 2020, 691, 108513. [Google Scholar] [CrossRef]
- Andérica-Romero, A.C.; Hernández-Damián, J.; Vázquez-Cervantes, G.I.; Torres, I.; Pedraza-Chaverri, J. The MLN4924 inhibitor exerts a neuroprotective effect against oxidative stress injury via Nrf2 protein accumulation. Redox Biol. 2016, 8, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Luo, H.; Chang, L.; Wang, S.; Geng, X.; Kang, L.; Zhong, Y.; Cao, Y.; Wang, R.; Yang, X.; et al. The NEDD8-activating enzyme inhibitor MLN4924 reduces ischemic brain injury in mice. Proc. Natl. Acad. Sci. USA 2022, 119, e2111896119. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Cui, J.; Zhao, F.; Yang, L.; Xu, X.; Shi, Y.; Wei, B. Cardioprotective effect of MLN4924 on ameliorating autophagic flux impairment in myocardial ischemia-reperfusion injury by Sirt1. Redox Biol. 2021, 46, 102114. [Google Scholar] [CrossRef] [PubMed]
- Swords, R.T.; Erba, H.P.; DeAngelo, D.J.; Bixby, D.L.; Altman, J.K.; Maris, M.; Hua, Z.; Blakemore, S.J.; Faessel, H.; Sedarati, F.; et al. Pevonedistat (MLN 4924), a First-in-Class NEDD 8-activating enzyme inhibitor, in patients with acute myeloid leukaemia and myelodysplastic syndromes: A phase 1 study. Br. J. Haematoloy 2015, 169, 534–543. [Google Scholar] [CrossRef]
- Mao, H.; Tang, Z.; Li, H.; Sun, B.; Tan, M.; Fan, S.; Zhu, Y.; Sun, Y. Neddylation inhibitor MLN4924 suppresses cilia formation by modulating AKT1. Protein Cell 2019, 10, 726–744. [Google Scholar] [CrossRef] [PubMed]
- Xiong, C.; Zhou, L.; Tan, J.; Song, S.; Bao, X.; Zhang, N.; Ding, H.; Zhao, J.; He, J.-X.; Miao, Z.-H. Development of potent NEDD8-activating enzyme inhibitors bearing a pyrimidotriazole scaffold. J. Med. Chem. 2021, 64, 6161–6178. [Google Scholar] [CrossRef]
- Salaroglio, I.C.; Belisario, D.C.; Bironzo, P.; Ananthanarayanan, P.; Ricci, L.; Digiovanni, S.; Fontana, S.; Napoli, F.; Sandri, A.; Facolmatà, C.; et al. SKP2 drives the sensitivity to neddylation inhibitors and cisplatin in malignant pleural mesothelioma. J. Expreimental Clin. Cancer Res. 2022, 41, 1–19. [Google Scholar] [CrossRef]
- Nawrocki, S.T.; Kelly, K.R.; Smith, P.G.; Keaton, M.; Carraway, H.; Sekeres, M.A.; Maciejewski, J.P.; Carew, J.S. The NEDD8-activating enzyme inhibitor MLN4924 disrupts nucleotide metabolism and augments the efficacy of cytarabine. Clin. Cancer Res. 2015, 21, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Megger, D.A.; Abou-Eid, S.; Zülch, B.; Sitek, B. Systematic analysis of synergistic proteome modulations in a drug combination of cisplatin and MLN4924. Mol. Omics 2018, 14, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Czuczman, N.M.; Barth, M.J.; Dwar, R.; Mavis, C.; Klener, P., Jr.; Czuczman, M.S.; Hernandez-Ilizaliturri, F.J. Evaluation of the Anti-Tumor Activity of MLN4924, A Novel NEDD8 Activating Enzyme Inhibitor, in Pre-Clinical Models of Rituximab Chemotherapy-Sensitive or-Resistant B-Cell Lymphoma. Blood 2012, 120, 2761. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-González, A.; Ramírez-Díaz, I.; Guzmán-Linares, J.; Sarvari, P.; Sarvari, P.; Rubio, K. ncRNAs Orchestrate Chemosensitivity Induction by Neddylation Blockades. Cancers 2024, 16, 825. https://doi.org/10.3390/cancers16040825
Pérez-González A, Ramírez-Díaz I, Guzmán-Linares J, Sarvari P, Sarvari P, Rubio K. ncRNAs Orchestrate Chemosensitivity Induction by Neddylation Blockades. Cancers. 2024; 16(4):825. https://doi.org/10.3390/cancers16040825
Chicago/Turabian StylePérez-González, Andrea, Ivonne Ramírez-Díaz, Josué Guzmán-Linares, Pouya Sarvari, Pourya Sarvari, and Karla Rubio. 2024. "ncRNAs Orchestrate Chemosensitivity Induction by Neddylation Blockades" Cancers 16, no. 4: 825. https://doi.org/10.3390/cancers16040825
APA StylePérez-González, A., Ramírez-Díaz, I., Guzmán-Linares, J., Sarvari, P., Sarvari, P., & Rubio, K. (2024). ncRNAs Orchestrate Chemosensitivity Induction by Neddylation Blockades. Cancers, 16(4), 825. https://doi.org/10.3390/cancers16040825





