Statistical Analysis of the Role of Cavity Flexibility in Thermostability of Proteins
Abstract
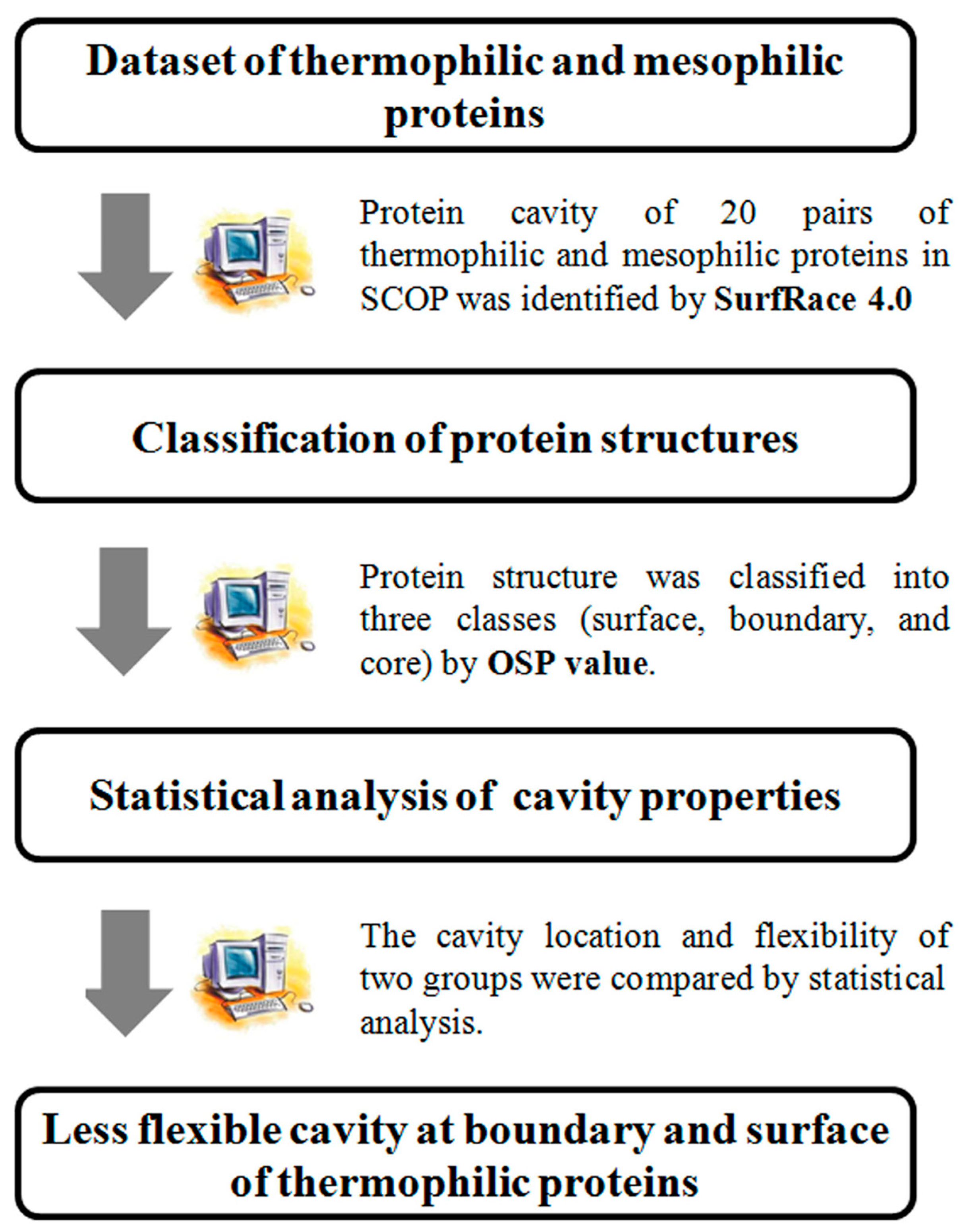
:1. Introduction
2. Materials and Methods
2.1. Dataset of Homologous Thermophilic and Mesophilic Proteins
2.2. Structure Classification by Residual Packing Value
2.3. Calculation of Cavity Flexibility
2.4. Statistical Analysis
3. Results and Discussion
3.1. Comparison of Cavity Properties in Thermophilic and Mesophilic Proteins
3.2. Difference in Cavity Location and Flexibility between Thermophilic and Mesophilic Proteins
3.3. Examples of Engineering the Flexible Cavities in the Surface Areas to Improve Protein Thermostability
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Zamani, R.; Rahpeyma, S.S.; Aliakbari, M.; Naderi, M.; Yazdanei, M.; Aminzadeh, S.; Khezri, J.; Haghbeen, K.; Karkhane, A.A. Enhancing the Thermostability of Cellulase from Clostridium thermocellum via Salt Bridge Interactions. Biotechnol. Bioprocess Eng. 2023, 28, 684–694. [Google Scholar] [CrossRef]
- Kim, H.; Lee, U.-J.; Lim, G.-M.; Kim, J.-Y.; Lee, J.; Song, H.; Kim, E.-J.; Kim, J.; Hwang, N.S.; Kim, B.-G. Stability Enhancement of Target Enzymes via Tyrosinase-Mediated Site-Specific Polysaccharide Coating. Biotechnol. Bioprocess Eng. 2023, 28, 862–873. [Google Scholar] [CrossRef]
- Chen, L.; Jiang, K.; Zhou, Y.; Zhu, L.; Chen, X. Improving the Thermostability of α-Glucosidase from Xanthomonas campestris through Proline Substitutions Guided by Semi-rational Design. Biotechnol. Bioprocess Eng. 2022, 27, 631–639. [Google Scholar] [CrossRef]
- Kumar, S.; Tsai, C.J.; Nussinov, R. Factors enhancing protein thermostability. Protein Eng. 2000, 13, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Pack, S.P.; Yoo, Y.J. Packing-based difference of structural features between thermophilic and mesophilic proteins. Int. J. Biol. Macromol. 2005, 35, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Arnold, F.H.; Wintrode, P.L.; Miyazaki, K.; Gershenson, A. How enzymes adapt: Lessons from directed evolution. Trends Biochem. Sci. 2001, 26, 100–106. [Google Scholar] [CrossRef]
- Eijsink, V.G.H.; Bjørk, A.; Gåseidnes, S.; Sirevåg, R.; Synstad, B.; Burg, B.V.D.; Vriend, G. Rational engineering of enzyme stability. J. Biotechnol. 2004, 113, 105–120. [Google Scholar] [CrossRef]
- Abraham, T.; Pack, S.P.; Je Yoo, Y. Stabilization of Bacillus subtilis Lipase A by increasing the residual packing. Biocatal. Biotransformation 2005, 23, 217–224. [Google Scholar] [CrossRef]
- Dubey, V.K.; Jagannadham, M.V. Roles for cavities in protein structure: New insights. Curr. Proteom. 2008, 5, 157–160. [Google Scholar] [CrossRef]
- Frieden, C. Effects of point mutations in a hinge region on the stability, folding, and enzymatic activity of Escherichia coli dihydrofolate reductase. Biochemistry 1991, 30, 7801–7809. [Google Scholar]
- Glyakina, A.V.; Garbuzynskiy, S.O.; Lobanov, M.Y.; Galzitskaya, O.V. Different packing of external residues can explain differences in the thermostability of proteins from thermophilic and mesophilic organisms. Bioinformatics 2007, 23, 2231–2238. [Google Scholar] [CrossRef] [PubMed]
- Gåseidnes, S.; Synstad, B.; Jia, X.; Kjellesvik, H.; Vriend, G.; Eijsink, V.G.H. Stabilization of a chitinase from Serratia marcescens by Gly→Ala and Xxx→Pro mutations. Protein Eng. 2003, 16, 841–846. [Google Scholar] [CrossRef] [PubMed]
- Sonavane, S.; Chakrabarti, P. Cavities and atomic packing in protein structures and interfaces. PLoS Comput. Biol. 2008, 4, e1000188. [Google Scholar] [CrossRef] [PubMed]
- Karshikoff, A.; Ladenstein, R. Proteins from thermophilic and mesophilic organisms essentially do not differ in packing. Protein Eng. 1998, 11, 867–872. [Google Scholar] [CrossRef] [PubMed]
- Szilágyi, A.; Závodszky, P. Structural differences between mesophilic, moderately thermophilic and extremely thermophilic protein subunits: Results of a comprehensive survey. Structure 2000, 8, 493–504. [Google Scholar] [CrossRef] [PubMed]
- Pattabiraman, N.; Ward, K.B.; Fleming, P.J. Occluded molecular surface: Analysis of protein packing. J. Mol. Recognit. 1995, 8, 334–344. [Google Scholar] [CrossRef]
- Yokota, K.; Satou, K.; Ohki, S.y. Comparative analysis of protein thermostability: Differences in amino acid content and substitution at the surfaces and in the core regions of thermophilic and mesophilic proteins. Sci. Technol. Adv. Mater. 2006, 7, 255–262. [Google Scholar] [CrossRef]
- Joo, J.C.; Pack, S.P.; Kim, Y.H.; Yoo, Y.J. Thermostabilization of Bacillus circulans xylanase: Computational optimization of unstable residues based on thermal fluctuation analysis. J. Biotechnol. 2011, 151, 56–65. [Google Scholar] [CrossRef]
- Fleming, P.J.; Richards, F.M. Protein packing: Dependence on protein size, secondary structure and amino acid composition. J. Mol. Biol. 2000, 299, 487–498. [Google Scholar] [CrossRef]
- Tsodikov, O.V.; Thomas Record Jr, M.; Sergeev, Y.V. Novel computer program for fast exact calculation of accessible and molecular surface areas and average surface curvature. J. Comput. Chem. 2002, 23, 600–609. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, T.; Chen, K.; Shen, S.; Ruan, J.; Kurgan, L. On the relation between residue flexibility and local solvent accessibility in proteins. Proteins: Struct. Funct. Bioformatics 2009, 76, 617–636. [Google Scholar] [CrossRef] [PubMed]
- Mendenhall, W.; Beaver, R.J. Introduction to Probability and Statistics, 8th ed.; PWS-KENT Publish: Boston, MA, USA, 1991; pp. 322–391. [Google Scholar]
- Agarwal, R.; Shrestha, U.R.; Chu, X.-Q.; Petridis, L.; Smith, J.C. Mesophilic Pyrophosphatase Function at High Temperature: A Molecular Dynamics Simulation Study. Biophys. J. 2020, 119, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Kalimeri, M.; Rahaman, O.; Melchionna, S.; Sterpone, F. How Conformational Flexibility Stabilizes the Hyperthermophilic Elongation Factor G-Domain. J. Phys. Chem. B 2013, 117, 13775–13785. [Google Scholar] [CrossRef] [PubMed]
- Bogin, O.; Levin, I.; Hacham, Y.; Tel-Or, S.; Peretz, M.; Frolow, F.; Burstein, Y. Structural basis for the enhanced thermal stability of alcohol dehydrogenase mutants from the mesophilic bacterium Clostridium beijerinckii: Contribution of salt bridging. Protein Sci. 2002, 11, 2561–2574. [Google Scholar] [CrossRef] [PubMed]
- Joo, J.C.; Pohkrel, S.; Pack, S.P.; Yoo, Y.J. Thermostabilization of Bacillus circulans xylanase via computational design of a flexible surface cavity. J. Biotechnol. 2010, 146, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Kuhlman, B.; Baker, D. Native protein sequences are close to optimal for their structures. Proc. Natl. Acad. Sci. USA 2000, 97, 10383–10388. [Google Scholar] [CrossRef]
- Guo, R.; Cang, Z.; Yao, J.; Kim, M.; Deans, E.; Wei, G.; Kang, S.-g.; Hong, H. Structural cavities are critical to balancing stability and activity of a membrane-integral enzyme. Proc. Natl. Acad. Sci. USA 2020, 117, 22146–22156. [Google Scholar] [CrossRef]
- Min, K.; Kim, H.; Park, H.J.; Lee, S.; Jung, Y.J.; Yoon, J.H.; Lee, J.-S.; Park, K.; Yoo, Y.J.; Joo, J.C. Improving the catalytic performance of xylanase from Bacillus circulans through structure-based rational design. Bioresour. Technol. 2021, 340, 125737. [Google Scholar] [CrossRef]
- Min, K.; Kim, H.T.; Park, S.J.; Lee, S.; Jung, Y.J.; Lee, J.-S.; Yoo, Y.J.; Joo, J.C. Improving the organic solvent resistance of lipase a from Bacillus subtilis in water–ethanol solvent through rational surface engineering. Bioresour. Technol. 2021, 337, 125394. [Google Scholar] [CrossRef]
- Jun, C.; Joo, J.C.; Lee, J.H.; Kim, Y.H. Thermostabilization of glutamate decarboxylase B from Escherichia coli by structure-guided design of its pH-responsive N-terminal interdomain. J. Biotechnol. 2014, 174, 22–28. [Google Scholar] [CrossRef]
- Goldenzweig, A.; Fleishman, S.J. Principles of Protein Stability and Their Application in Computational Design. Annu. Rev. Biochem. 2018, 87, 105–129. [Google Scholar] [CrossRef] [PubMed]
- Tran, K.-N.T.; Kumaravel, A.; Hong, S.H. Impact of the Synthetic Scaffold Strategy on the Metabolic Pathway Engineering. Biotechnol. Bioprocess Eng. 2023, 28, 379–385. [Google Scholar] [CrossRef]
- Matthay, P.; Schalck, T.; Verstraeten, N.; Michiels, J. Strategies to Enhance the Biosynthesis of Monounsaturated Fatty Acids in Escherichia coli. Biotechnol. Bioprocess Eng. 2023, 28, 36–50. [Google Scholar] [CrossRef]
- Woo, J.-M.; Kang, Y.-S.; Lee, S.-M.; Park, S.; Park, J.-B. Substrate-binding Site Engineering of Candida antarctica Lipase B to Improve Selectivity for Synthesis of 1-monoacyl-sn-glycerols. Biotechnol. Bioprocess Eng. 2022, 27, 234–243. [Google Scholar] [CrossRef]
- Khobragade, T.P.; Pagar, A.D.; Giri, P.; Sarak, S.; Jeon, H.; Joo, S.; Goh, Y.; Park, B.-S.; Yun, H. Biocatalytic Cascade for Synthesis of Sitagliptin Intermediate Employing Coupled Transaminase. Biotechnol. Bioprocess Eng. 2023, 28, 300–309. [Google Scholar] [CrossRef]
- Kang, H.; Sriramulu, D.K.; Lee, S.-G. In silico Study on Binding Specificities of Cellular Retinol Binding Protein and Its Q108R Mutant. Biotechnol. Bioprocess Eng. 2022, 27, 126–134. [Google Scholar] [CrossRef]



| SCOP Fold Name | Thermophilic Proteins | Mesophilic Proteins | ||
|---|---|---|---|---|
| PDB | Organism | PDB | Organism | |
| Adenine nucleotide alpha hydrolase-like | 1V8FA | Thermus thermophilus HB8 | 1IHOB | Escherichia coli |
| Amionoacid dehydrogenase-like, N-terminal domain | 1EUZA | Thermococcus profundus | 1HRDA | Clostridium symbiosum |
| ATC-like | 1ML4A | Pyrococcus abyssi | 1D09A | Escherichia coli |
| Chorismate synthase, AroC | 1Q1LA | Aquifex aeolicus | 1QXOC | Streptococcus pneumoniae |
| DHS-like NAD/FAD-binding domain | 1ICIB | Archaeoglobus fulgidus | 1S5PA | Escherichia coli |
| Ferredoxin-like | 1MROD | Methanothermobacter marburgensis | 1E6YA | Methanosarcina barkeri |
| GroES-like | 1RJWA | Geobacillus stearothermophilus | 1LLUA | Pseudomonas aeruginosa |
| HAD-like | 1F5SA | Methanococcus jannaschii | 1NNLB | Homo sapiens |
| LDH C-terminal domain like | 1IZ9A | Thermus thermophilus | 1B8PA | Aquaspirillum arcticum |
| Macrodomain-like | 1VHUA | Archaeoglobus fulgidus | 1SPVA | Escherichia coli |
| NagB/RpiA/CoA transferase | 1LK7A | Pyrococcus horikoshii | 1M0SA | Haemophilus influenzae |
| Nucleotide-diphospho-sugar transferase | 1LVWC | Methanothermobacter thermautotrophicus | 1IIMA | Salmonella enterica |
| Phosphoglycerate kinase | 1PHP | Geobacillus stearothermophilus | 1VJCA | Sus scrofa |
| PLP-depedent transferase | 1KL1A | Geobacillus stearothermophilus | 1DFOA | Escherichia coli |
| S-adenosyl-L-methionine-depedent methyltransferase | 1JQ3A | Thermotoga maritima | 1IY9A | Bacillus subtilis |
| Subtilisin-like | 1THM | Thermoactinomyces vulgaris | 1NDOA | Bacillus subtilis |
| Thiamin diphosphate-binding fold (THDP-binding) | 1UMCB | Thermus thermophilus | 1OLSB | Homo sapiens |
| Thiolase-like | 1J3NA | Thermus thermophilus | 1IY9A | Streptococcus pneumoniae |
| YebC-like | 1LFPA | Aquifex aeolicus | 1KONA | Escherichia coli |
| YrdC/RibB | 1PVWA | Methanocaldococcus jannaschii | 1K4PA | Magnaporthe grisea |
| Df | t0.1 | t0.05 | t0.025 | t0.01 | t0.005 |
|---|---|---|---|---|---|
| Inf (>30) | 1.282 | 1.645 | 1.960 | 2.326 | 2.576 |
| Proteins | Number of Proteins | Number of Cavities (Total a/Average per Protein b/Average per Residue c) | Average Volume of Cavity (Å3) | Average Surface Area of Cavity (Å2) |
|---|---|---|---|---|
| Thermophilic proteins | 20 | 369/18.45/0.056 | 26.65 | 45.53 |
| Mesophilic proteins | 20 | 355/17.75/0.056 | 27.51 | 46.20 |
| Structure Index (OSP Value) | Frequency | ||||||
|---|---|---|---|---|---|---|---|
| Thermo | SD a | Flexibility b | Meso | SD a | Flexibility b | t-test c | |
| Surface (0.00~0.250) | 0.0081 | ±0.0202 | −0.0034 | 0.0091 | ±0.0280 | 0.1985 | −0.1233 |
| Boundary (0.250~0.500) | 0.7253 | ±0.1546 | −0.2428 | 0.8025 | ±0.1203 | −0.2047 | −1.7621 |
| Core (0.500~0.750) | 0.2673 | ±0.1573 | −0.6484 | 0.1884 | ±0.1099 | −0.5111 | 1.8386 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, S.Y.; Yoon, J.; An, Y.J.; Lee, S.; Cha, H.-G.; Pandey, A.; Yoo, Y.J.; Joo, J.C. Statistical Analysis of the Role of Cavity Flexibility in Thermostability of Proteins. Polymers 2024, 16, 291. https://doi.org/10.3390/polym16020291
Hong SY, Yoon J, An YJ, Lee S, Cha H-G, Pandey A, Yoo YJ, Joo JC. Statistical Analysis of the Role of Cavity Flexibility in Thermostability of Proteins. Polymers. 2024; 16(2):291. https://doi.org/10.3390/polym16020291
Chicago/Turabian StyleHong, So Yeon, Jihyun Yoon, Young Joo An, Siseon Lee, Haeng-Geun Cha, Ashutosh Pandey, Young Je Yoo, and Jeong Chan Joo. 2024. "Statistical Analysis of the Role of Cavity Flexibility in Thermostability of Proteins" Polymers 16, no. 2: 291. https://doi.org/10.3390/polym16020291
APA StyleHong, S. Y., Yoon, J., An, Y. J., Lee, S., Cha, H. -G., Pandey, A., Yoo, Y. J., & Joo, J. C. (2024). Statistical Analysis of the Role of Cavity Flexibility in Thermostability of Proteins. Polymers, 16(2), 291. https://doi.org/10.3390/polym16020291










