An Optimized Tobacco Hairy Root Induction System for Functional Analysis of Nicotine Biosynthesis-Related Genes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Culture Conditions
2.2. R. rhizogenes Strains and Hairy Root Induction
2.3. Expression of Foreign Genes in Tobacco Hairy Root
2.4. Transgenic Hairy Root Verification
2.5. RNA Extraction and Quantitative RT-PCR
2.6. Determination of Nicotine Content
3. Statistical Analysis
4. Results
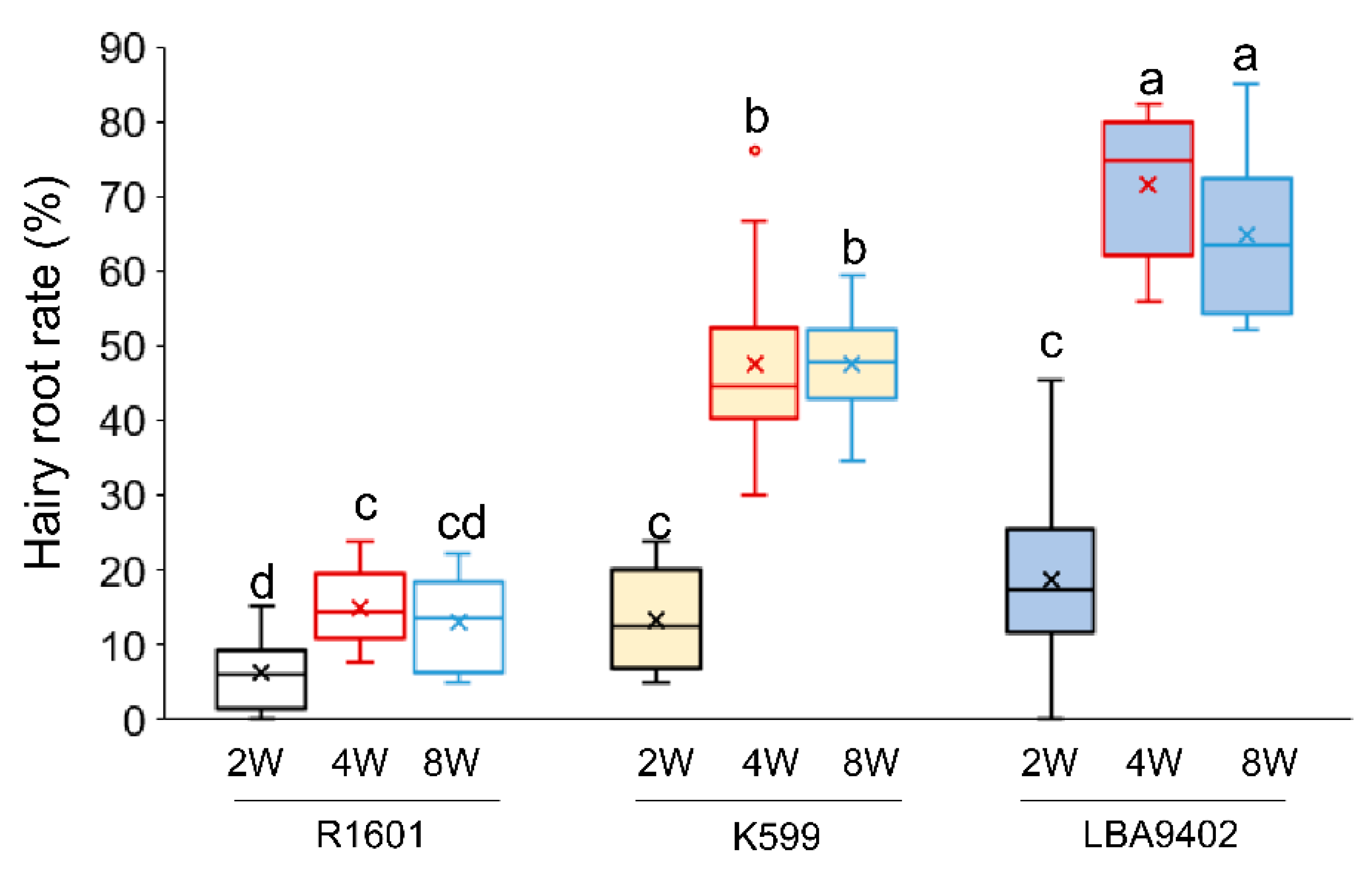
4.1. Effects of the Explant Age and R. rhizogenes Strains on Hairy Root Induction
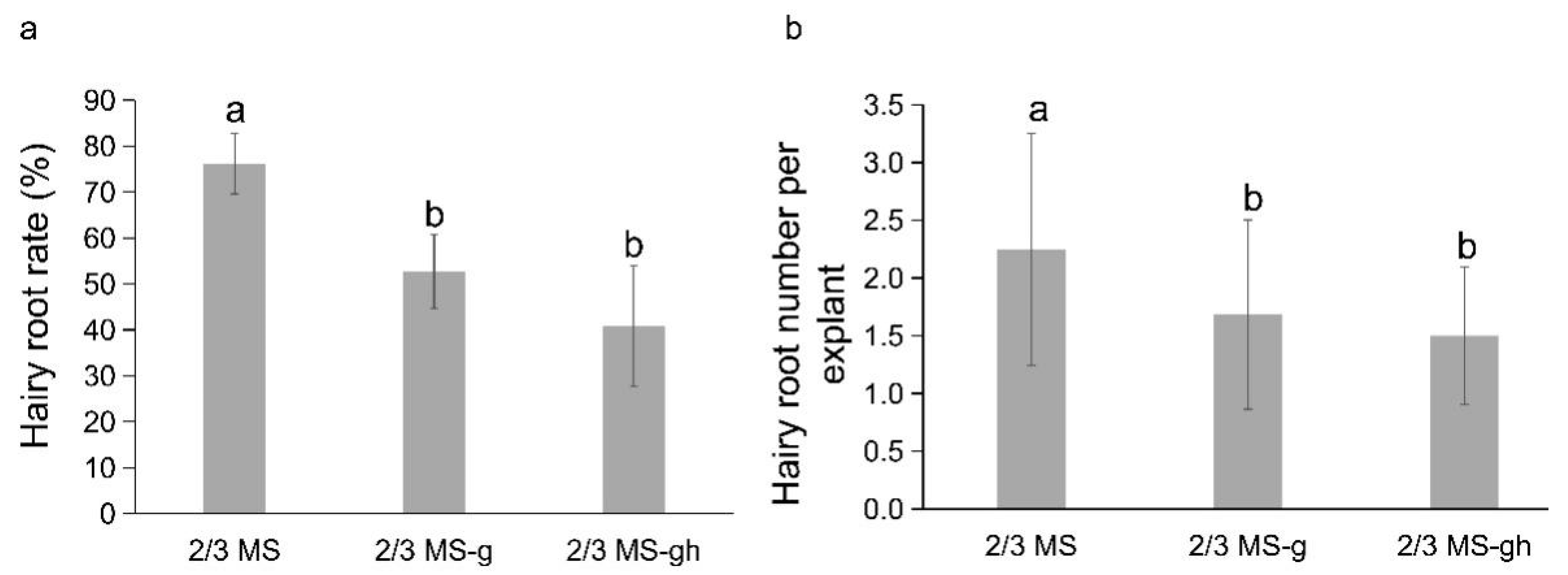
4.2. Effects of Medium Additives on Hairy Roots Induction
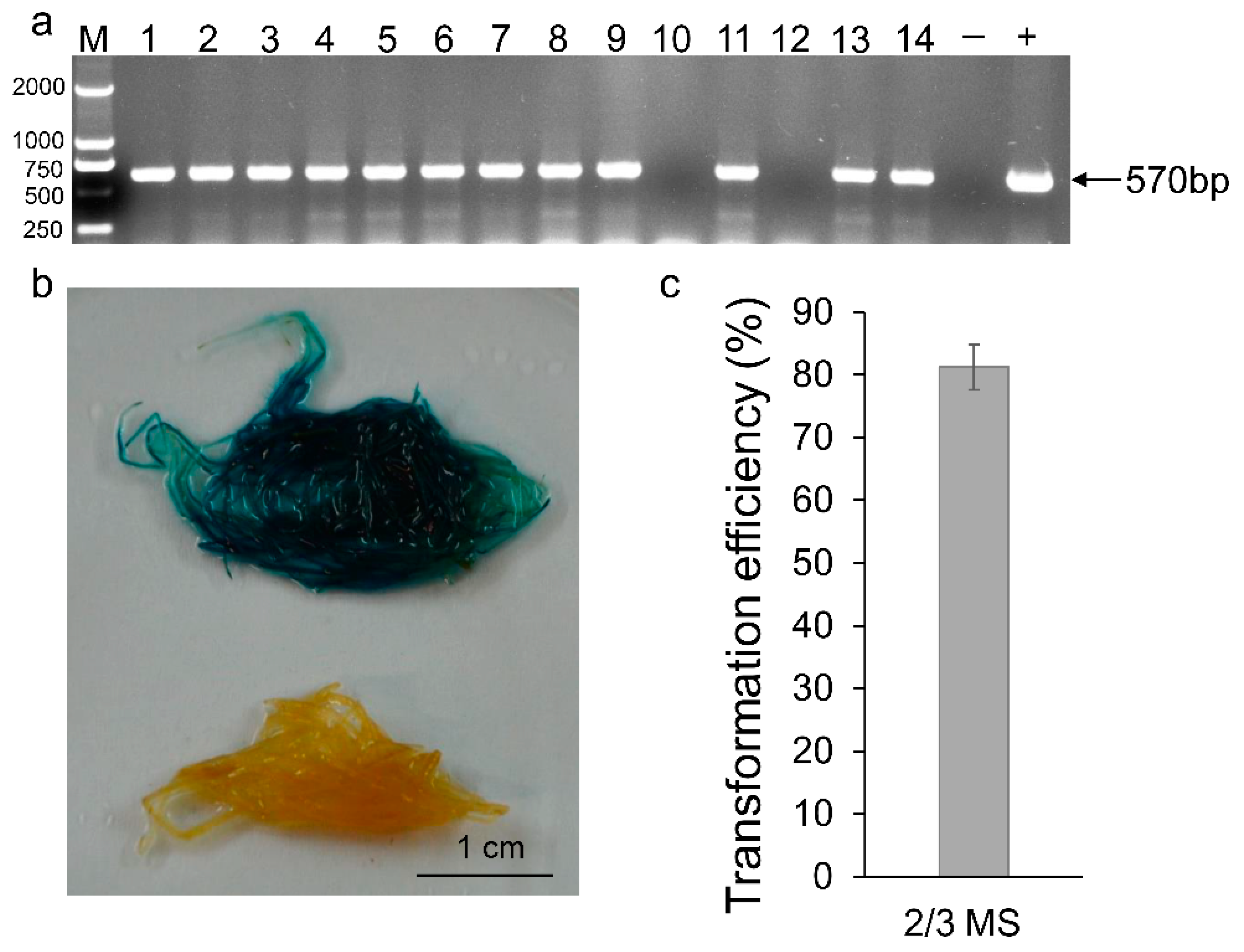
4.3. R. rhizogenes Strain LBA9402 as a Cargo for Foreign Gene Delivery and Expression in Hairy Root
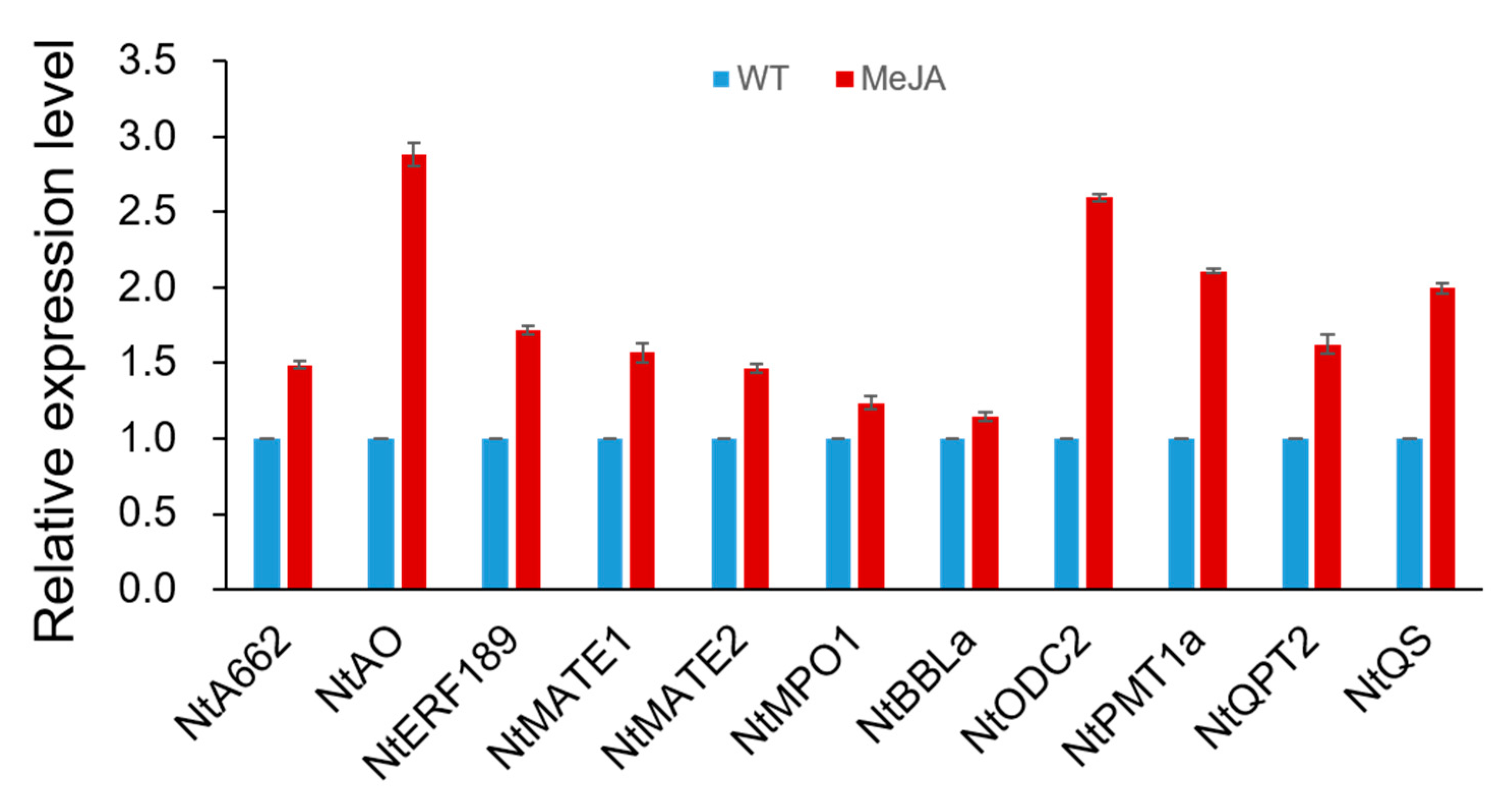
4.4. Expression of Genes Related to Nicotine Biosynthesis in Hairy Roots
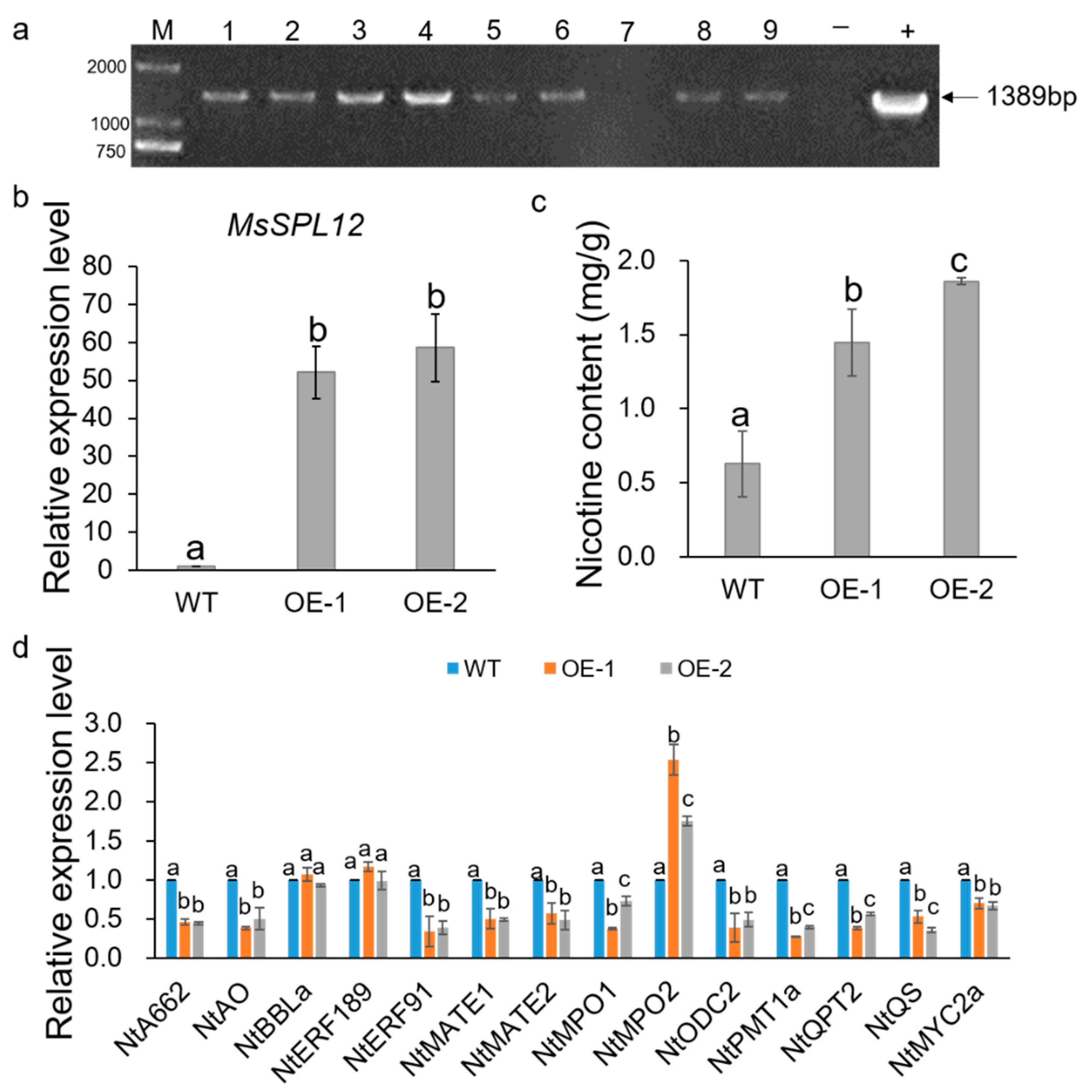
4.5. Effects of Overexpression of MsSPL12 on Nicotine Biosynthesis in Tobacco Hairy Roots
5. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Flores-Felix, J.D.; Menendez, E.; Peix, A.; Garcia-Fraile, P.; Velazquez, E. History and current taxonomic status of genus Agrobacterium. Syst Appl. Microbiol. 2020, 43, 126046. [Google Scholar] [CrossRef] [PubMed]
- Wallaart, T.E.; Pras, N.; Quax, W.J. Isolation and identification of dihydroartemisinic acid hydroperoxide from Artemisia annua: A novel biosynthetic precursor of Artemisinin. J. Nat. Prod. 1999, 62, 1160–1162. [Google Scholar] [CrossRef] [PubMed]
- Chahardoli, M.; Fazeli, A.; Ghabooli, M. Recombinant production of bovine lactoferrin-derived antimicrobial peptide in tobacco hairy roots expression system. Plant Physiol. Bioch. 2018, 123, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.L.; Zhu, X.; Shao, J.; Wu, Y.; Tang, Y. Transcriptional response of the catharanthine biosynthesis pathway to methyl jasmonate/nitric oxide elicitation in Catharanthus roseus hairy root culture. Appl. Microbiol. Biot. 2010, 88, 737–750. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Padh, H.; Shrivastava, N. Hairy root cultures: A suitable biological system for studying secondary metabolic pathways in plants. Eng. Life Sci. 2013, 13, 62–75. [Google Scholar] [CrossRef]
- Hou, X.J.; Li, J.M.; Liu, B.L.; Wei, L. Co-expression of basic helix–loop–helix protein (bHLH) and transcriptional activator-Myb genes induced anthocyanin biosynthesis in hairy root culture of Nicotiana tabacum L. and Ipomea tricolor. Acta. Physiol. Plant 2017, 39, 59. [Google Scholar] [CrossRef]
- Hidalgo, D.; Georgiev, M.; Marchev, A.; Bru-Martínez, R.; Cusido, R.M.; Corchete, P.; Palazon, J. Tailoring tobacco hairy root metabolism for the production of stilbenes. Sci. Rep. 2017, 7, 17976. [Google Scholar] [CrossRef]
- Alderete, L.G.S.; Guido, M.E.; Agostini, E.; Mas, P. Identification and characterization of key circadian clock genes of tobacco hairy roots: Putative regulatory role in xenobiotic metabolism. Environ Sci. Pollut. R. 2018, 25, 1597–1608. [Google Scholar] [CrossRef] [Green Version]
- Daspute, A.A.; Yunxuan, X.; Gu, M.; Kobaysshi, Y.; Wagh, S.; Panche, A.; Koyama, H. Agrobacterium rhizogenes -mediated hairy roots transformation as a tool for exploring aluminum-responsive genes function. Future Sci. OA 2019, 5, 3. [Google Scholar] [CrossRef] [Green Version]
- Pilaisangsuree, V.; Anuwan, P.; Supdensong, K.; Lumpa, P.; Kongbangkerd, A.; Limmongkon, A. Enhancement of adaptive response in peanut hairy root by exogenous signalling molecules under cadmium stress. J. Plant Physiol. 2020, 254, 153278. [Google Scholar] [CrossRef]
- Zipfel, C. Pattern-recognization receptors in plant innate immunity. Curr. Opin. Immunol. 2008, 20, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Dewey, R.E.; Boss, W.; Phillippy, B.Q.; Qu, R.D. Enhanced Agrobacterium-mediated transformation effificiencies in monocot cells is associated with attenuated defense responses. Plant Mol. Biol. 2013, 81, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Thwe, A.; Arasu, M.V.; Li, X.; Park, C.H.; Kim, S.J.; Al-Dhabi, N.A.; Park, S.U. Effect of different Agrobacterium rhizogenes strains on hairy root induction and phenylpropanoid biosynthesis in tartary buckwheat (Fagopyrum tataricum Gaertn). Front Microbiol. 2016, 7, 318. [Google Scholar] [CrossRef] [PubMed]
- Park, C.H.; Zhao, S.; Yeo, H.J.; Park, Y.E.; Baska, T.B.; Arasu, M.V.; Al-Dhabi, N.A.; Park, S.U. Comparison of different strains of Agrobacterium rhizogenes for hairy root induction and betulin and betulinic acid production in Morusalba. Nat. Prod. Commun. 2017, 12, 1934578X1701200403. [Google Scholar] [CrossRef] [Green Version]
- Surendra, K.; Kakoli, B. A comparative study of hairy root culture induction efficiency in four medicinally important plants using Agrobacterium rhizogenes. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 625–633. [Google Scholar]
- Le Flem-Bonhomme, V.; Laurain-Mattar, D.; Fliniaux, M.A. Hairy root induction of Papaver somniferum var. album, a difficult-to-transform plant, by A. rhizogenes LBA 9402. Planta 2004, 218, 890–893. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Wang, X.; Cao, L.; Ji, J.; Liu, T.; Duan, K. Highly efficient Agrobacterium rhizogenes-mediated hairy root transformation for gene functional and gene editing analysis in soybean. Plant Methods 2021, 17, 73. [Google Scholar] [CrossRef]
- Rana, M.M.; Han, Z.X.; Song, D.P.; Liu, G.F.; Li, D.X.; Wan, X.C.; Karthikeyan, A.; Wei, S. Effect of medium supplements on Agrobacterium rhizogenes mediated hairy root induction from the callus tissues of camellia sinensis var. sinensis. Int. J. Mol. Sci. 2016, 17, 1132. [Google Scholar] [CrossRef] [Green Version]
- Saunders, J.A. Investigations of vacuoles isolated from tobacco: I. Quantitation of nicotine. Plant Physiol. 1979, 64, 74–78. [Google Scholar] [CrossRef] [Green Version]
- Baldwin, I.T. Mechanism of damage-induced alkaloid production in wild tobacco. J. Chem. Ecol. 1989, 15, 1661–1680. [Google Scholar] [CrossRef]
- Sui, X.; He, X.; Song, Z.; Gao, Y.; Zhao, L.; Jiao, F.; Kong, G.; Li, Y.; Han, S.; Wang, B. The gene NtMYC2a acts as a‘master switch’in the regulation of JA-induced nicotine accumulation in tobacco. Plant Biol. 2021, 23, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Afrin, S.; Huang, J.J.; Luo, Z.Y. JA-mediated transcriptional regulation of secondary metabolism in medicinal plants. Sci. Bull. 2015, 60, 1062. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Wang, D.D.; Fang, X.; Chen, X.; Mao, Y. Plant specialized metabolism regulated by jasmonate signaling. Plant Cell Physiol. 2019, 60, 2638–2647. [Google Scholar] [CrossRef] [PubMed]
- Shoji, T.; Hashimoto, T. Expression of a tobacco nicotine biosynthesis gene depends on the JRE4 transcription factor in heterogenous tomato. J. Plant Res. 2019, 132, 173–180. [Google Scholar] [CrossRef]
- Cao, Y.; Chen, R.; Wang, W.; Gao, X. SmSPL6 induces phenolic acid biosynthesis and affects root development in Salvia miltiorrhiza. Int. J. Mol. Sci. 2021, 22, 7895. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.B.; Liu, Y.Q.; Chen, D.Y.; Chen, F.Y.; Fang, X.; Hong, G.J.; Wang, L.J.; Wang, J.W.; Chen, X.Y. Jasmonate response decay and defense metabolite accumulation contributes to age-regulated dynamics of plant insect resistance. Nat. Commun. 2017, 8, 13925. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Kan, Y.; Liu, W. Differential expression of miRNAs in response to topping in flue-cured tobacco (Nicotiana tabacum) roots. PLoS ONE 2011, 6, e28565. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Liu, Y.; Teng, F.; Cen, H.; Yan, J.; Lin, S.; Li, D.; Zhang, W. Heterogeneous expression of Osa-MIR156bc increases abiotic stress resistance and forage quality of alfalfa. Crop J. 2021, 9, 1135–1144. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Kim, J.S.; Lee, S.Y.; Park, S.U. Resveratrol production in hairy root culture of peanut, Arachis hypogaea L. transformed with different Agrobacterium rhizogenes strains. Afr. J. Biotechnol. 2008, 7, 3788–3790. [Google Scholar]
- Cao, D.; Hou, W.; Song, S.; Sun, H.; Wu, C.; Gao, Y.; Han, T. Assessment of conditions affecting Agrobacterium rhizogenes-mediated transformation of soybean. Plant Cell. Tiss. Org. 2009, 96, 45–52. [Google Scholar] [CrossRef]
- An, G.; Ebert, P.; Mitra, A.; Ha, S.B. Binary vectors. In Plant Molecular Biology Manual; Gelvin, S.B., Schilperoort, R.A., Eds.; Kluwer Academic: Dordrecht, The Netherlands, 1988; pp. 1–19. [Google Scholar]
- Xiao, H.; Wang, Y.; Liu, D.F.; Wang, W.M.; Li, X.; Zhao, X.; Xu, J.; Zhai, W.; Zhu, L. Functional analysis of the rice AP3 homologue OsMADS16 by RNA interference. Plant Mol. Biol. 2003, 52, 957–966. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cen, H.; Yan, J.; Zhang, Y.; Zhang, W. Inside out: High-efficiency plant regeneration and Agrobacterium-mediated transformation of upland and lowland switchgrass cultivars. Plant Cell Rep. 2015, 34, 1099–1108. [Google Scholar] [CrossRef] [PubMed]
- Lalitha, S. Primer premier 5. Biotech Softw. Internet Rep. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- Jefferson, R. Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Bio. Rep. 1987, 5, 387–405. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Sui, X.; Zhang, H.; Song, Z.; Gao, Y.; Li, W.; Li, M.; Zhao, L.; Li, Y.; Wang, B. Ethylene response factor NtERF91 positively regulates alkaloid accumulations in tobacco (Nicotiana tabacum L.). Bioch. Bioph. Res. Commun. 2019, 517, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Setamam, N.M.; Sidik, N.J.; Rahman, Z.A.; Zain, C.R.C.M. Induction of hairy roots by various strains of Agrobacterium rhizogenes in different types of Capsicum species explants. BMC Res. 2014, 7, 414–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Cao, Y.; Wang, Y.; Fu, C.; Dai, S. Agrobacterium rhizogenes-mediated transformation system of Spinacia oleracea. Chin. Bull. Bot. 2019, 4, 515–521. [Google Scholar]
- Veena, V.; Taylor, C.G. Agrobacterium rhizogenes: Recent developments and promising applications. In Vitro Cell. Dev.-Plant 2007, 43, 383–403. [Google Scholar] [CrossRef]
- Shanks, J.V.; Morgan, J. Plant ‘hairy root’ culture. Curr. Opin. Biotech. 1999, 10, 151–155. [Google Scholar] [CrossRef]
- Zhou, M.L.; Zhu, X.M.; Shao, J.R.; Tang, Y.X.; Wu, Y.M. Production and metabolic engineering of bioactive substances in plant hairy root culture. Appl. Microbiol. Biot. 2011, 90, 1229–1239. [Google Scholar] [CrossRef] [PubMed]
- Sudo, H.; Yamakawa, T.; Yamazaki, M.; Aimi, N.; Saito, K. Bioreactor production of camptothecin by hairy root cultures of Ophiorrhiza pumila. Biotechnol. Lett. 2002, 24, 359–363. [Google Scholar] [CrossRef]
- Geyter, N.D.; Gholami, A.; Goormachtig, S.; Goossens, A. Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci. 2012, 17, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, I.T.; Zhang, Z.P.; Diab, N.; Ohnmeiss, T.E.; McCloud, E.S.; Lynds, G.Y.; Schmelz, L.E. Quantification, correlations and manipulations of wound-induced changes in jasmonic acid and nicotine in Nicotiana sylvestris. Planta 1997, 201, 397–404. [Google Scholar] [CrossRef]
- Dewey, R.E.; Xie, J. Molecular genetics of alkaloid biosynthesis in nicotiana tabacum. Phytochemistry 2013, 94, 10–27. [Google Scholar] [CrossRef]
- Kajikawa, M.; Sierro, N.; Kawaguchi, H.; Bakaher, N.; Ivanov, N.V.; Hashimoto, T.; Shoji, T. Genomic insights into the evolution of the nicotine biosynthesis pathway in tobacco. Plant Physiol. 2017, 174, 999–1011. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Wang, H.Y. The miR156/SPL module, a regulatory hub and versatile toolbox, gears up crops for enhanced agronomic traits. Mol. Plant 2015, 8, 677–688. [Google Scholar] [CrossRef] [Green Version]
- Li, X.Y.; Hou, Y.M.; Xie, X.; Li, H.; Li, X.; Zhu, Y.; Zhai, L.; Zhang, C.; Bian, S. A blueberry MIR156a-SPL12 module coordinates the accumulation of chlorophylls and anthocyanins during fruit ripening. J. Exp. Bot. 2020, 71, 5976–5989. [Google Scholar] [CrossRef]
- Lv, Z.Y.; Wang, Y.; Liu, Y.; Peng, B.W.; Zhang, L.; Tang, K.X.; Chen, W.S. The SPB-box transcription factor AaSPL2 positively regulates artemisinin biosynthesis in Artemisia annua L. Front. Plant Sci. 2019, 10, 409. [Google Scholar] [CrossRef] [Green Version]
- Katoh, A.; Shoji, T.; Hashimoto, T. Molecular cloning of N-methylputrescine oxidase from tobacco. Plant Cell Physiol. 2007, 48, 550–554. [Google Scholar] [CrossRef] [PubMed]
- Kajikawa, M.; Shoji, T.; Kato, A.; Hashimoto, T. Vacuole-localized berberine bridge enzyme-like proteins are required for a late step of nicotine biosynthesis in tobacco. Plant Physiol. 2011, 155, 2010–2022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shoji, T.; Kajikawa, M.; Hashimoto, T. Clustered transcription factor genes regulate nicotine biosynthesis in tobacco. Plant Cell 2010, 22, 3390–3409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B. Factors in Nicotine Biosynthesis in Tobacco. Ph.D. Thesis, North Carolina State University, Raleigh, NC, USA, 2011. [Google Scholar]
- Chen, H.X.; Wang, B.W.; Geng, S.S.; Arellano, C.; Chen, S.; Qu, R. Effects of overexpression of jasmonic acid biosynthesis genes on nicotine accumulation in tobacco. Plant Direct. 2018, 1, e00036. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, S.; Liu, Y.; Yan, J.; Lin, S.; Zhang, W.; Wang, B. An Optimized Tobacco Hairy Root Induction System for Functional Analysis of Nicotine Biosynthesis-Related Genes. Agronomy 2022, 12, 348. https://doi.org/10.3390/agronomy12020348
Qin S, Liu Y, Yan J, Lin S, Zhang W, Wang B. An Optimized Tobacco Hairy Root Induction System for Functional Analysis of Nicotine Biosynthesis-Related Genes. Agronomy. 2022; 12(2):348. https://doi.org/10.3390/agronomy12020348
Chicago/Turabian StyleQin, Shangqian, Yanrong Liu, Jianping Yan, Shiwen Lin, Wanjun Zhang, and Bingwu Wang. 2022. "An Optimized Tobacco Hairy Root Induction System for Functional Analysis of Nicotine Biosynthesis-Related Genes" Agronomy 12, no. 2: 348. https://doi.org/10.3390/agronomy12020348






