Cells 2022, 11(19), 3151; https://doi.org/10.3390/cells11193151 - 7 Oct 2022
Cited by 3 | Viewed by 2970
| Correction
Abstract
Among the 33 human adhesion G-protein-coupled receptors (aGPCRs), a unique subfamily of GPCRs, only ADGRF4, encoding GPR115, shows an obvious skin-dominated transcriptomic profile, but its expression and function in skin is largely unknown. Here, we report that GPR115 is present in a
[...] Read more.
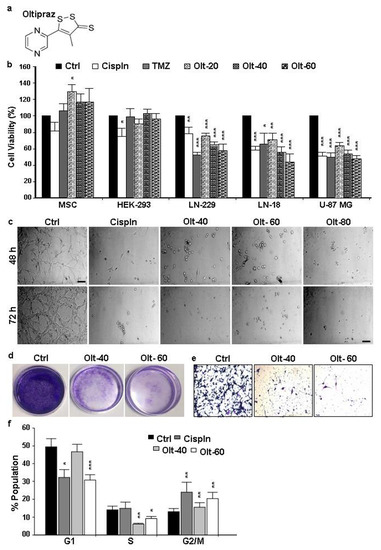
Among the 33 human adhesion G-protein-coupled receptors (aGPCRs), a unique subfamily of GPCRs, only ADGRF4, encoding GPR115, shows an obvious skin-dominated transcriptomic profile, but its expression and function in skin is largely unknown. Here, we report that GPR115 is present in a small subset of basal and in most suprabasal, noncornified keratinocytes of the stratified epidermis, supporting epidermal transcriptomic data. In psoriatic skin, characterized by hyperproliferation and delayed differentiation, the expression of GPR115 and KRT1/10, the fundamental suprabasal keratin dimer, is delayed. The deletion of ADGRF4 in HaCaT keratinocytes grown in an organotypic mode abrogates KRT1 and reduces keratinocyte stratification, indicating a role of GPR115 in epidermal differentiation. Unexpectedly, endogenous GPR115, which is not glycosylated and is likely not proteolytically processed, localizes intracellularly along KRT1/10-positive keratin filaments in a regular pattern. Our data demonstrate a hitherto unknown function of GPR115 in the regulation of epidermal differentiation and KRT1.
Full article
(This article belongs to the Special Issue Structures, Regulation, Signaling, and Physiological Functions of Adhesion G-Protein Coupled Receptors)
►
Show Figures