Upregulated Immunogenic Cell-Death-Associated Gene Signature Predicts Reduced Responsiveness to Immune-Checkpoint-Blockade Therapy and Poor Prognosis in High-Grade Gliomas
Abstract
:1. Introduction
2. Material and Methods
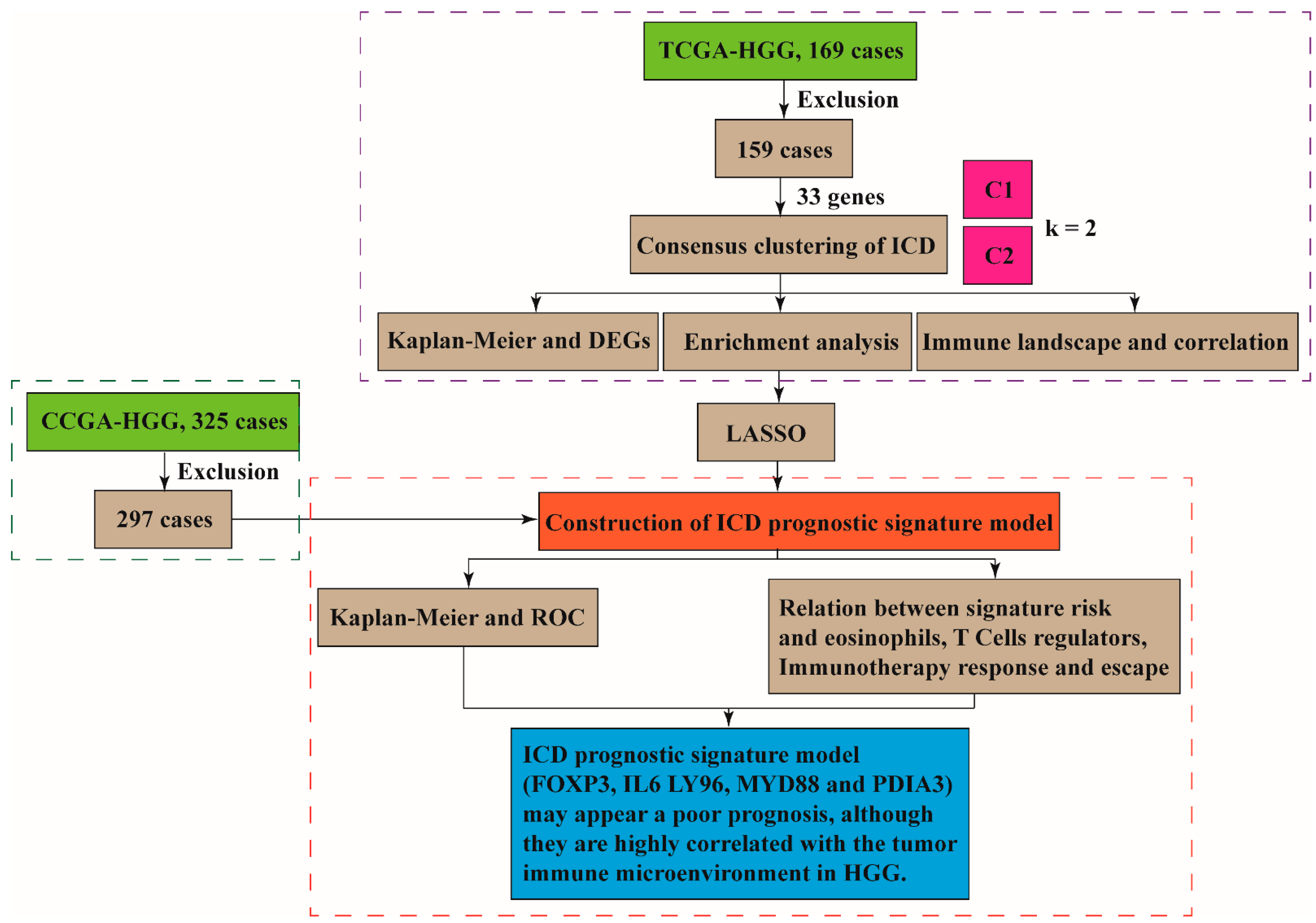
2.1. Datasets
2.2. Consensus Clustering of ICD in HGGs
2.3. Identification of Differentially Expressed Genes (DEGs), Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) [21,22]
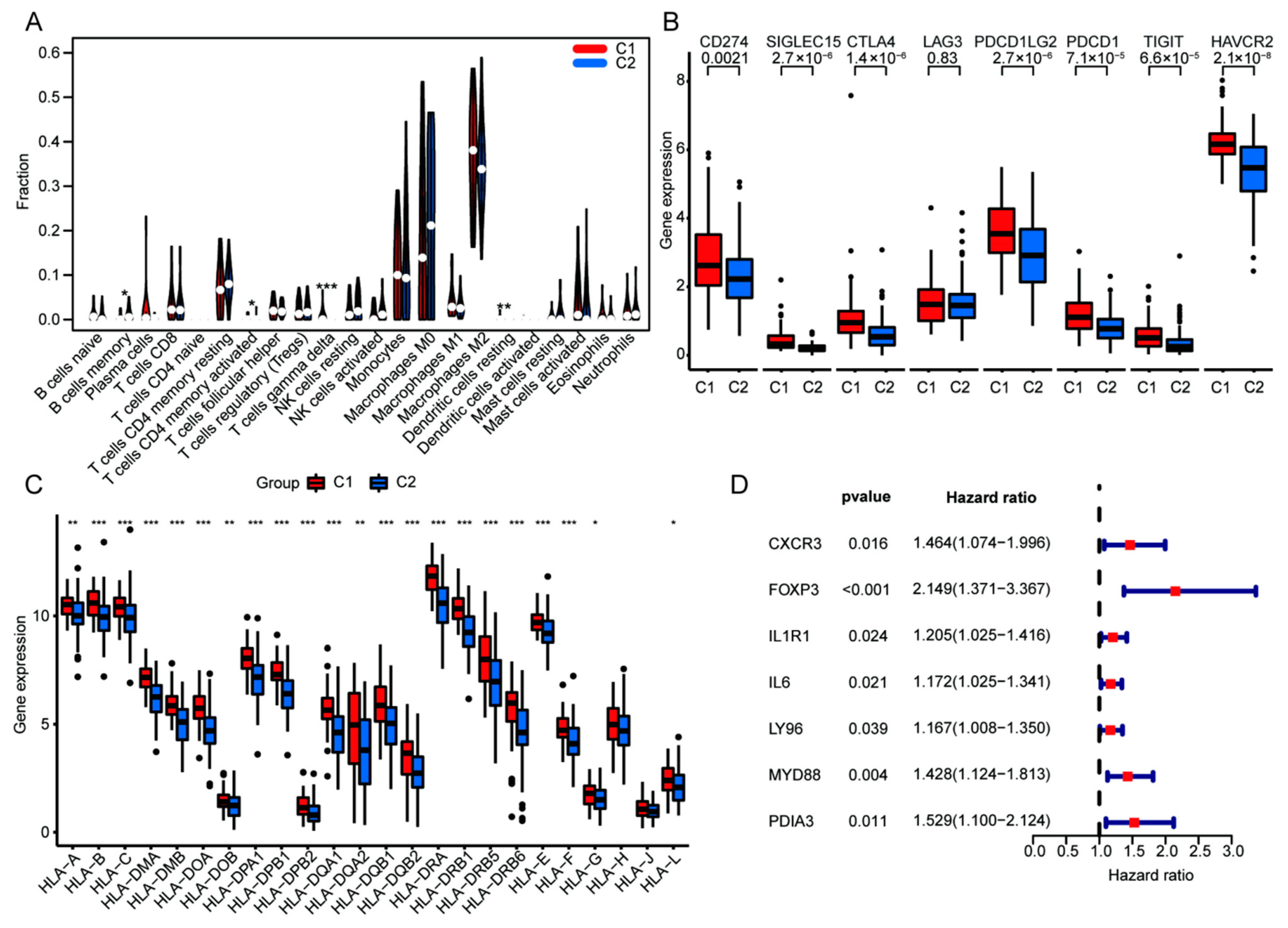
2.4. Characterization of Immune Landscape and Correlation [23]
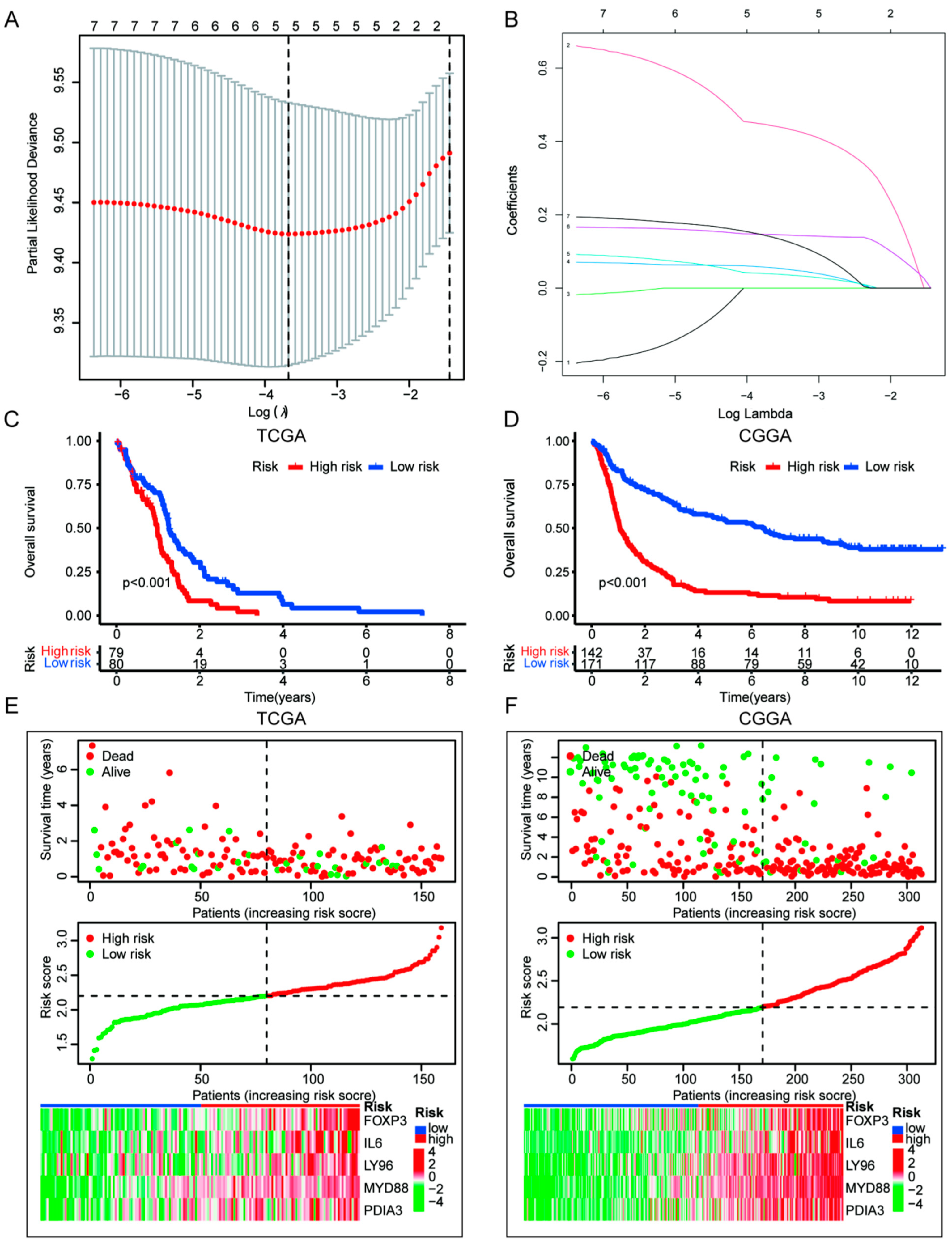
2.5. Construction of ICD Prognostic Signature Model and Survival Analysis [24,25]
2.6. Prediction of Response to ICB Immunotherapy [26,27]
2.7. Statistical Analysis
3. Results
3.1. The ICD-Associated Gene Signature in HGGs
3.2. Potential Biological Functions and Signal Pathways Associated with ICD Signature
3.3. Upregulated ICD Signature Was Correlated to Reduced Somatic Tumor Mutations and High Infiltration of Immune Cells in HGGs
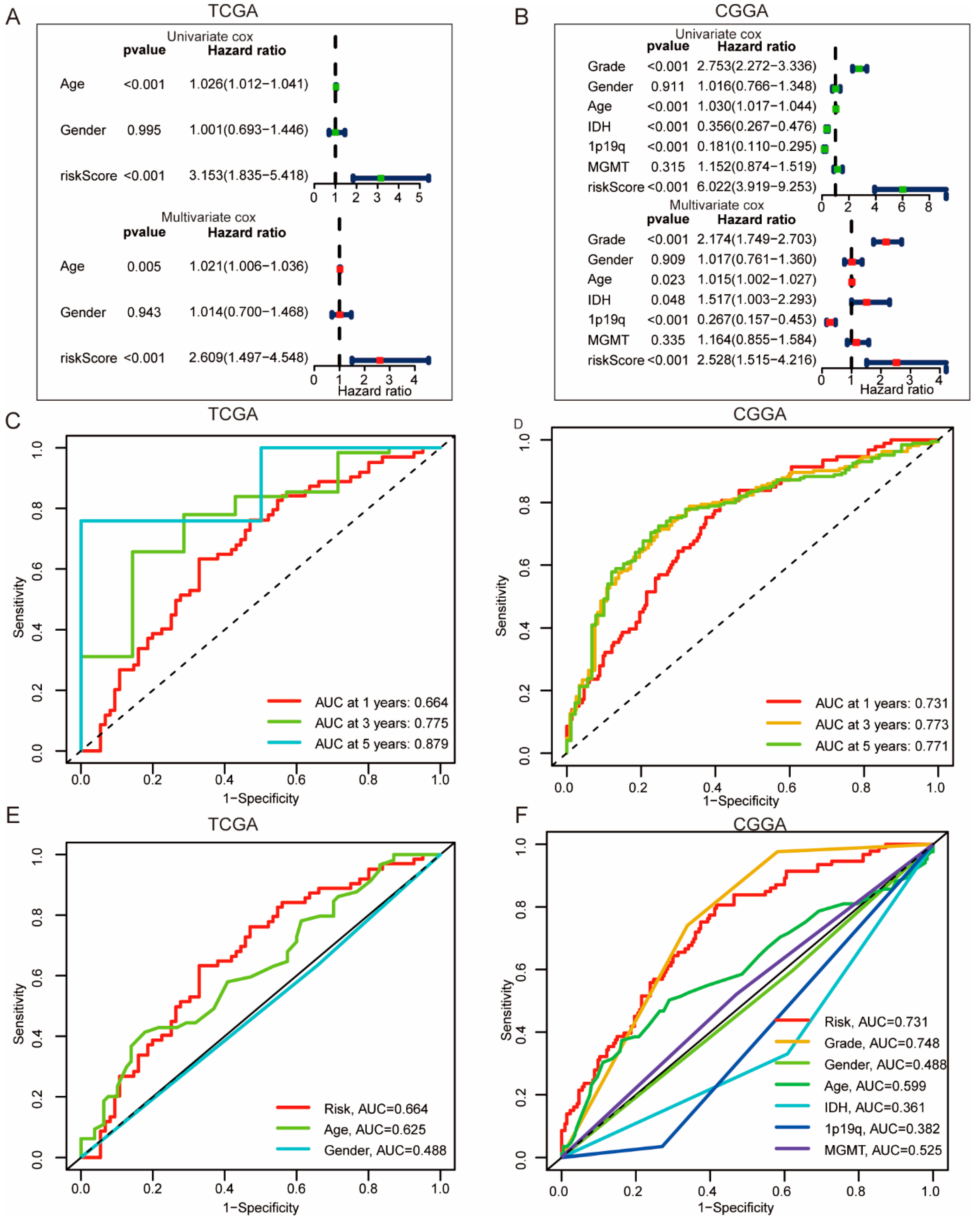
3.4. Construction of ICD Risk Signature Model and Its Value in HGGs Patient Prognosis Prediction
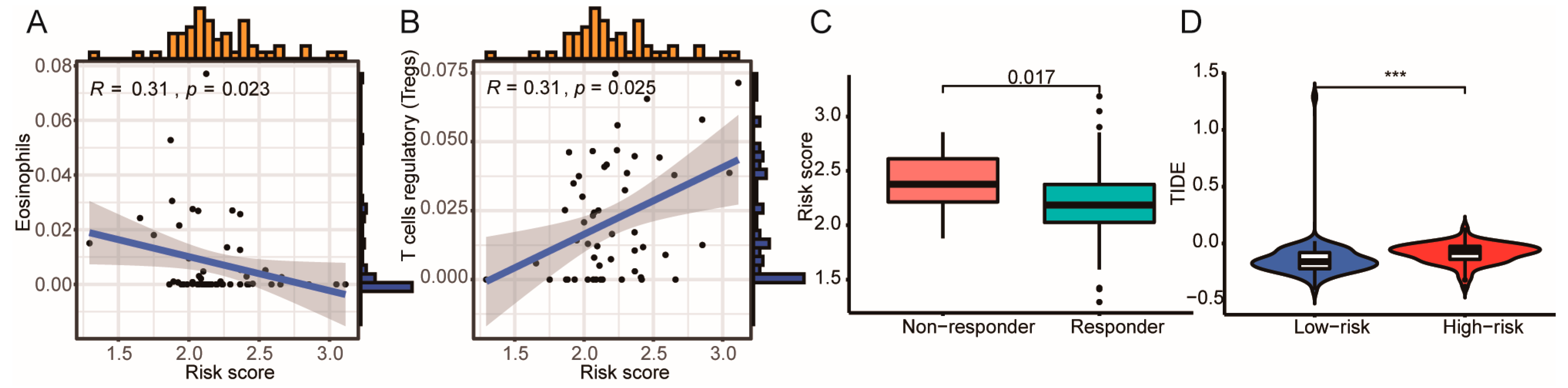
3.5. High ICD Gene Signature Is Associated with Reduced ICB Immunotherapy Response
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Ethics Statement
Acknowledgments
Conflicts of Interest
References
- Senhaji, N.; Squalli Houssaini, A.; Lamrabet, S.; Louati, S.; Bennis, S. Molecular and circulating biomarkers in patients with glioblastoma. Int. J. Mol. Sci. 2022, 23, 7474. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; He, M.Z.; Li, T.; Yang, X. MRI combined with PET-CT of different tracers to improve the accuracy of glioma diagnosis: A systematic review and meta-analysis. Neurosurg. Rev. 2019, 42, 185–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reiss, S.N.; Yerram, P.; Modelevsky, L.; Grommes, C. Retrospective review of safety and efficacy of programmed cell death-1 inhibitors in refractory high grade gliomas. J. Immunother. Cancer 2017, 5, 99. [Google Scholar] [CrossRef]
- Venkataramani, V.; Yang, Y.; Schubert, M.C.; Reyhan, E.; Tetzlaff, S.K.; Wißmann, N.; Botz, M.; Soyka, S.J.; Beretta, C.A.; Pramatarov, R.L.; et al. Glioblastoma hijacks neuronal mechanisms for brain invasion. Cell 2022, 185, 2899–2917. [Google Scholar] [CrossRef]
- Miyauchi, J.T.; Tsirka, S.E. Advances in immunotherapeutic research for glioma therapy. J. Neurol. 2018, 265, 741–756. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Vitale, I.; Warren, S.; Adjemian, S.; Agostinis, P.; Martinez, A.B.; Chan, T.A.; Coukos, G.; Demaria, S.; Deutsch, E.; et al. Consensus guidelines for the definition, detection and interpretation of immunogenic cell death. J. Immunother. Cancer 2020, 8, e000337. [Google Scholar] [CrossRef] [Green Version]
- Yatim, N.; Cullen, S.; Albert, M.L. Dying cells actively regulate adaptive immune responses. Nat. Rev. Immunol. 2017, 17, 262–275. [Google Scholar] [CrossRef]
- Fucikova, J.; Kepp, O.; Kasikova, L.; Petroni, G.; Yamazaki, T.; Liu, P.; Zhao, L.; Spisek, R.; Kroemer, G.; Galluzzi, L. Detection of immunogenic cell death and its relevance for cancer therapy. Cell Death Dis. 2020, 11, 1013. [Google Scholar] [CrossRef]
- Deng, H.; Yang, W.; Zhou, Z.; Tian, R.; Lin, L.; Ma, Y.; Song, J.; Chen, X. Targeted scavenging of extracellular ROS relieves suppressive immunogenic cell death. Nat. Commun. 2020, 11, 4951. [Google Scholar] [CrossRef]
- Ladoire, S.; Enot, D.; Andre, F.; Zitvogel, L.; Kroemer, G. Immunogenic cell death-related biomarkers: Impact on the survival of breast cancer patients after adjuvant chemotherapy. Oncoimmunology 2015, 5, e1082706. [Google Scholar] [CrossRef]
- Soh, J.; Hamada, A.; Fujino, T.; Mitsudomi, T. Perioperative therapy for non-small cell lung cancer with immune checkpoint inhibitors. Cancers 2021, 13, 4035. [Google Scholar] [CrossRef] [PubMed]
- Fucikova, J.; Moserova, I.; Urbanova, L.; Bezu, L.; Kepp, O.; Cremer, I.; Salek, C.; Strnad, P.; Kroemer, G.; Galluzzi, L.; et al. Prognostic and predictive value of DAMPs and DAMP-associated processes in cancer. Front. Immunol. 2015, 6, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Yang, J.; Luo, L.; Jiang, M.; Qin, B.; Yin, H.; Zhu, C.; Yuan, X.; Zhang, J.; Luo, Z.; et al. Targeting photodynamic and photothermal therapy to the endoplasmic reticulum enhances immunogenic cancer cell death. Nat. Commun. 2019, 10, 3349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, A.; Tait, S.W.G. Targeting immunogenic cell death in cancer. Mol. Oncol. 2020, 14, 2994–3006. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yang, K.; Zhao, R.; Ji, T.; Wang, X.; Yang, X.; Zhang, Y.; Cheng, K.; Liu, S.; Hao, J.; et al. Inducing enhanced immunogenic cell death with nanocarrier-based drug delivery systems for pancreatic cancer therapy. Biomaterials 2016, 102, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Krysko, D.V.; Garg, A.D.; Kaczmarek, A.; Krysko, O.; Agostinis, P.; Vandenabeele, P. Immunogenic cell death and DAMPs in cancer therapy. Nat. Rev. Cancer 2012, 12, 860–875. [Google Scholar] [CrossRef]
- Garg, A.D.; De Ruysscher, D.; Agostinis, P. Immunological metagene signatures derived from immunogenic cancer cell death associate with improved survival of patients with lung, breast or ovarian malignancies: A large-scale meta-analysis. Oncoimmunology 2015, 5, e1069938. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wu, S.; Liu, F.; Ke, D.; Wang, X.; Pan, D.; Xu, W.; Zhou, L.; He, W. An immunogenic cell death-related classification predicts prognosis and response to immunotherapy in head and neck squamous cell carcinoma. Front. Immunol. 2021, 12, 781466. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, K.N.; Wang, Q.; Li, G.; Zeng, F.; Zhang, Y.; Wu, F.; Chai, R.; Wang, Z.; Zhang, C.; et al. Chinese Glioma Genome Atlas (CGGA): A comprehensive resource with functional genomic data from Chinese glioma patients. Genom. Proteom. Bioinform. 2021, 19, 1–12. [Google Scholar] [CrossRef]
- Wilkerson, M.D.; Hayes, D.N. ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics 2010, 26, 1572–1573. [Google Scholar] [CrossRef]
- Wang, H.; Zhu, H.; Zhu, W.; Xu, Y.; Wang, N.; Han, B.; Song, H.; Qiao, J. Bioinformatic analysis identifies potential key genes in the pathogenesis of Turner Syndrome. Front. Endocrinol. 2020, 11, 104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, D.D.; Li, J.D.; He, R.Q.; Li, M.X.; Pan, Y.Q.; He, X.L.; Dang, Y.W.; Chen, G. Highly expressed carbohydrate sulfotransferase 11 correlates with unfavorable prognosis and immune evasion of hepatocellular carcinoma. Cancer Med. 2022. online ahead of print. [Google Scholar] [CrossRef]
- Wei, N.; Gong, C.; Zhou, W.; Lei, Z.; Shi, Y.; Zhang, S.; Zhang, K.; Mao, Y.; Zhang, H. A ubiquitin-related gene signature for predicting prognosis and constructing molecular subtypes in osteosarcoma. Front. Pharmacol. 2022, 13, 904448. [Google Scholar] [CrossRef]
- Yin, T.; Yin, Y.; Qu, L. Exploration of shared gene signature with development of pre-eclampsia and cervical cancer. Front. Genet. 2022, 13, 972346. [Google Scholar] [CrossRef] [PubMed]
- Gately, L.; Collins, A.; Murphy, M.; Dowling, A. Age alone is not a predictor for survival in glioblastoma. J. Neurooncol. 2016, 129, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Guo, Z.; Yu, D.; Wang, Y.; Wang, Q.; Dong, Z.; Hu, W. A prognostic nomogram combining immune-related gene signature and clinical factors predicts survival in patients with lung adenocarcinoma. Front. Oncol. 2020, 10, 1300. [Google Scholar] [CrossRef]
- Wang, F.; Lin, H.; Su, Q.; Li, C. Cuproptosis-related lncRNA predict prognosis and immune response of lung adenocarcinoma. World J. Surg. Oncol. 2022, 20, 275. [Google Scholar] [CrossRef]
- Fu, J.; Li, K.; Zhang, W.; Wan, C.; Zhang, J.; Jiang, P.; Liu, X.S. Large-scale public data reuse to model immunotherapy response and resistance. Genome Med. 2020, 12, 21. [Google Scholar] [CrossRef] [Green Version]
- Jiang, P.; Gu, S.; Pan, D.; Fu, J.; Sahu, A.; Hu, X.; Li, Z.; Traugh, N.; Bu, X.; Li, B.; et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018, 24, 1550–1558. [Google Scholar] [CrossRef]
- Braunstein, S.; Raleigh, D.; Bindra, R.; Mueller, S.; Haas-Kogan, D. Pediatric high-grade glioma: Current molecular landscape and therapeutic approaches. J. Neurooncol. 2017, 134, 541–549. [Google Scholar] [CrossRef]
- Preusser, M.; Lim, M.; Hafler, D.A.; Reardon, D.A.; Sampson, J.H. Prospects of immune checkpoint modulators in the treatment of glioblastoma. Nat. Rev. Neurol. 2015, 11, 504–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, B.; Li, X.; Li, Y.; Zhang, J.; Zong, Z.; Zhang, H. Current immunotherapies for glioblastoma multiforme. Front. Immunol. 2021, 11, 603911. [Google Scholar] [CrossRef] [PubMed]
- Lim, M.; Xia, Y.; Bettegowda, C.; Weller, M. Current state of immunotherapy for glioblastoma. Nat. Rev. Clin. Oncol. 2018, 15, 422–442. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F.; Salinas, R.D.; Zhang, D.Y.; Nguyen, P.T.T.; Schnoll, J.G.; Wong, S.Z.H.; Thokala, R.; Sheikh, S.; Saxena, D.; Prokop, S.; et al. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell 2020, 180, 188–204.e22. [Google Scholar] [CrossRef]
- Turubanova, V.D.; Balalaeva, I.V.; Mishchenko, T.A.; Catanzaro, E.; Alzeibak, R.; Peskova, N.N.; Efimova, I.; Bachert, C.; Mitroshina, E.V.; Krysko, O.; et al. Immunogenic cell death induced by a new photodynamic therapy based on photosens and photodithazine. J. Immunother. Cancer 2019, 7, 350. [Google Scholar] [CrossRef]
- Xu, M.; Lu, J.H.; Zhong, Y.Z.; Jiang, J.; Shen, Y.Z.; Su, J.Y.; Lin, S.Y. Immunogenic cell death-relevant damage-associated molecular patterns and sensing receptors in triple-negative breast cancer molecular subtypes and implications for immunotherapy. Front Oncol. 2022, 12, 870914. [Google Scholar] [CrossRef]
- Dejaegher, J.; Solie, L.; Hunin, Z.; Sciot, R.; Capper, D.; Siewert, C.; Van Cauter, S.; Wilms, G.; van Loon, J.; Ectors, N.; et al. DNA methylation based glioblastoma subclassification is related to tumoral T-cell infiltration and patient survival. Neuro Oncol. 2021, 23, 240–250. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, C.; Wang, B.; Zhang, H.; Qin, G.; Li, C.; Cao, L.; Gao, Q.; Ping, Y.; Zhang, K.; et al. Regulatory T cells promote glioma cell stemness through TGF-β-NF-κB-IL6-STAT3 signaling. Cancer Immunol. Immunother. 2021, 70, 2601–2616. [Google Scholar] [CrossRef]
- Garg, A.D.; Dudek, A.M.; Ferreira, G.B.; Verfaillie, T.; Vandenabeele, P.; Krysko, D.V.; Mathieu, C.; Agostinis, P. ROS-induced autophagy in cancer cells assists in evasion from determinants of immunogenic cell death. Autophagy 2013, 9, 1292–1307. [Google Scholar] [CrossRef] [Green Version]
- Nie, K.; Li, J.; Peng, L.; Zhang, M.; Huang, W. Pan-cancer analysis of the characteristics of LY96 in prognosis and immunotherapy across human cancer. Front. Mol. Biosci. 2022, 9, 837393. [Google Scholar] [CrossRef]
- Billod, J.M.; Lacetera, A.; Guzmán-Caldentey, J.; Martín-Santamaría, S. Computational approaches to toll-like receptor 4 modulation. Molecules 2016, 21, 994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, N.; Xu, H.; Ou, Y.; Feng, Z.; Zhang, Q.; Zhu, Q.; Cai, Z. LPS-induced CXCR7 expression promotes gastric cancer proliferation and migration via the TLR4/MD-2 pathway. Diagn. Pathol. 2019, 14, 3. [Google Scholar] [CrossRef] [PubMed]
- Bi, F.; Wang, J.; Zheng, X.; Xiao, J.; Zhi, C.; Gu, J.; Zhang, Y.; Li, J.; Miao, Z.; Wang, Y.; et al. HSP60 participates in the anti-glioma effects of curcumin. Exp. Ther. Med. 2021, 21, 204. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Xiao, X.; Zhang, J. MYD88 is a potential prognostic gene and immune signature of tumor microenvironment for gliomas. Front. Oncol. 2021, 11, 654388. [Google Scholar] [CrossRef] [PubMed]
- Chiavari, M.; Ciotti, G.M.P.; Canonico, F.; Altieri, F.; Lacal, P.M.; Graziani, G.; Navarra, P.; Lisi, L. PDIA3 expression in glioblastoma modulates macrophage/microglia pro-tumor activation. Int. J. Mol. Sci. 2020, 21, 8214. [Google Scholar] [CrossRef]
- Tu, Z.; Ouyang, Q.; Long, X.; Wu, L.; Li, J.; Zhu, X.; Huang, K. Protein disulfide-isomerase A3 is a robust prognostic biomarker for cancers and predicts the immunotherapy response effectively. Front. Immunol. 2022, 13, 837512. [Google Scholar] [CrossRef]



| Dataset | TCGA (n = 159) | CGGA (n = 297) |
|---|---|---|
| Age (Years) | 59.41 ± 13.66 | 43.31 ± 12.00 |
| Gender (M/F) | 100/59 | 181/116 |
| Grade (2/3/4) | / | 91/73/133 |
| IDH mutation (Y/N) | / | 140/157 |
| 1p/19q co-deletion (Y/N) | / | 235/62 |
| MGMT promoter methylation (Y/N) | / | 152/145 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, X.; Guo, D.; Yang, X.; Chen, R.; Jiang, Q.; Zeng, Z.; Li, Y.; Li, Z. Upregulated Immunogenic Cell-Death-Associated Gene Signature Predicts Reduced Responsiveness to Immune-Checkpoint-Blockade Therapy and Poor Prognosis in High-Grade Gliomas. Cells 2022, 11, 3655. https://doi.org/10.3390/cells11223655
Tang X, Guo D, Yang X, Chen R, Jiang Q, Zeng Z, Li Y, Li Z. Upregulated Immunogenic Cell-Death-Associated Gene Signature Predicts Reduced Responsiveness to Immune-Checkpoint-Blockade Therapy and Poor Prognosis in High-Grade Gliomas. Cells. 2022; 11(22):3655. https://doi.org/10.3390/cells11223655
Chicago/Turabian StyleTang, Xin, Dongfang Guo, Xi Yang, Rui Chen, Qingming Jiang, Zhen Zeng, Yu Li, and Zhenyu Li. 2022. "Upregulated Immunogenic Cell-Death-Associated Gene Signature Predicts Reduced Responsiveness to Immune-Checkpoint-Blockade Therapy and Poor Prognosis in High-Grade Gliomas" Cells 11, no. 22: 3655. https://doi.org/10.3390/cells11223655
APA StyleTang, X., Guo, D., Yang, X., Chen, R., Jiang, Q., Zeng, Z., Li, Y., & Li, Z. (2022). Upregulated Immunogenic Cell-Death-Associated Gene Signature Predicts Reduced Responsiveness to Immune-Checkpoint-Blockade Therapy and Poor Prognosis in High-Grade Gliomas. Cells, 11(22), 3655. https://doi.org/10.3390/cells11223655








