Lagging Strand Initiation Processes in DNA Replication of Eukaryotes—Strings of Highly Coordinated Reactions Governed by Multiprotein Complexes
Abstract
:1. Introduction
2. Initiation of DNA Replication at Origins
3. Replication Forks
3.1. Leading Strand Synthesis
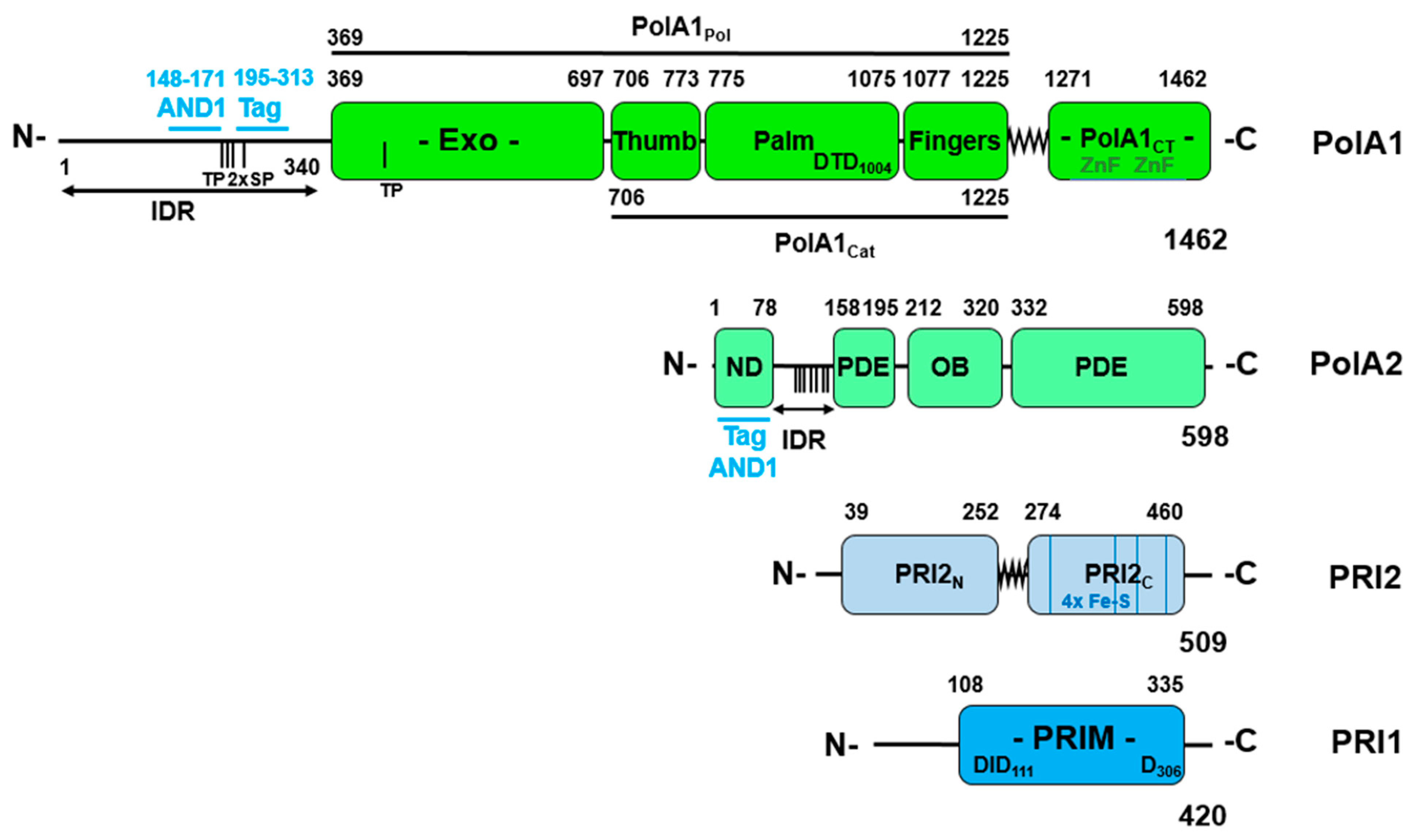
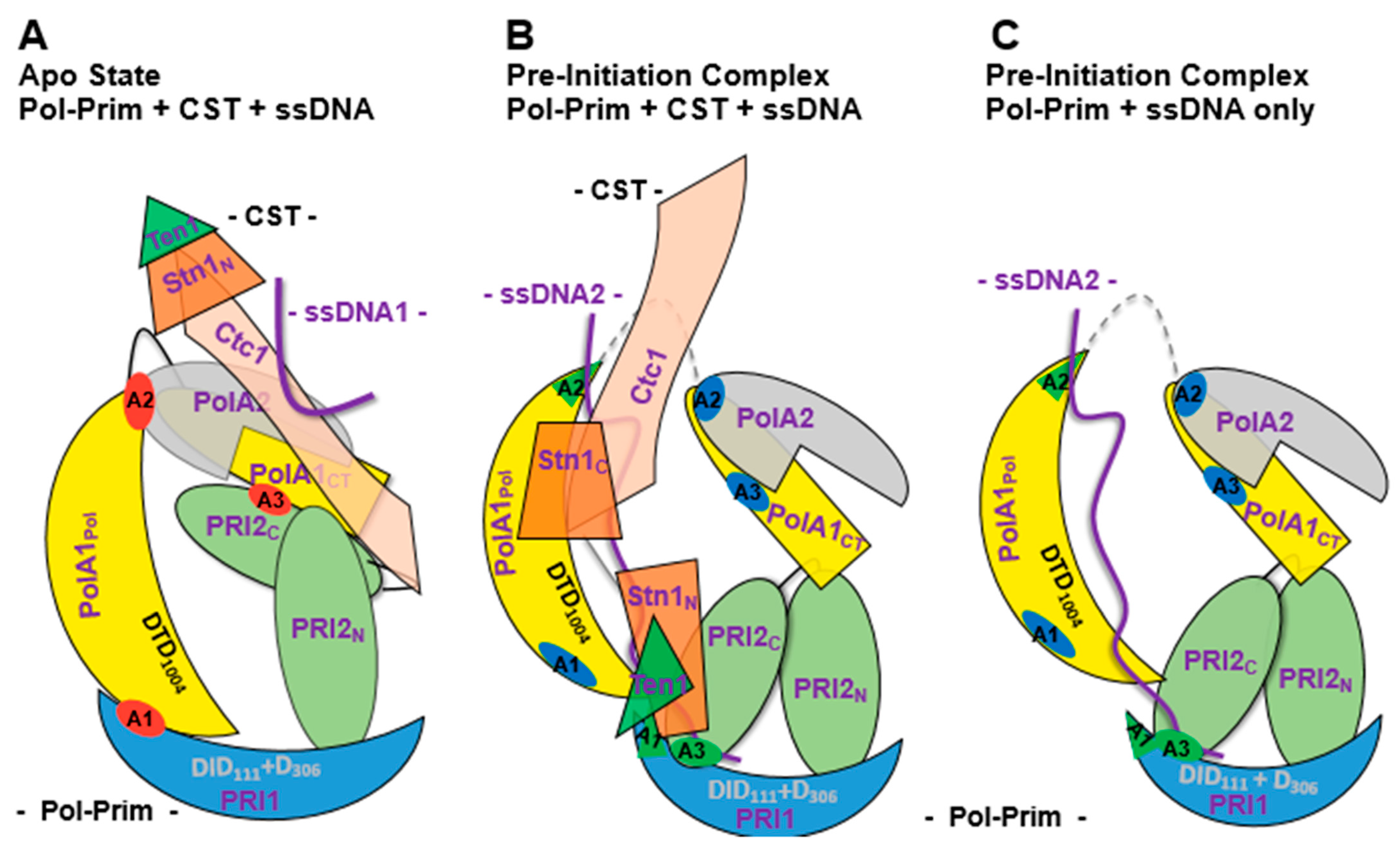
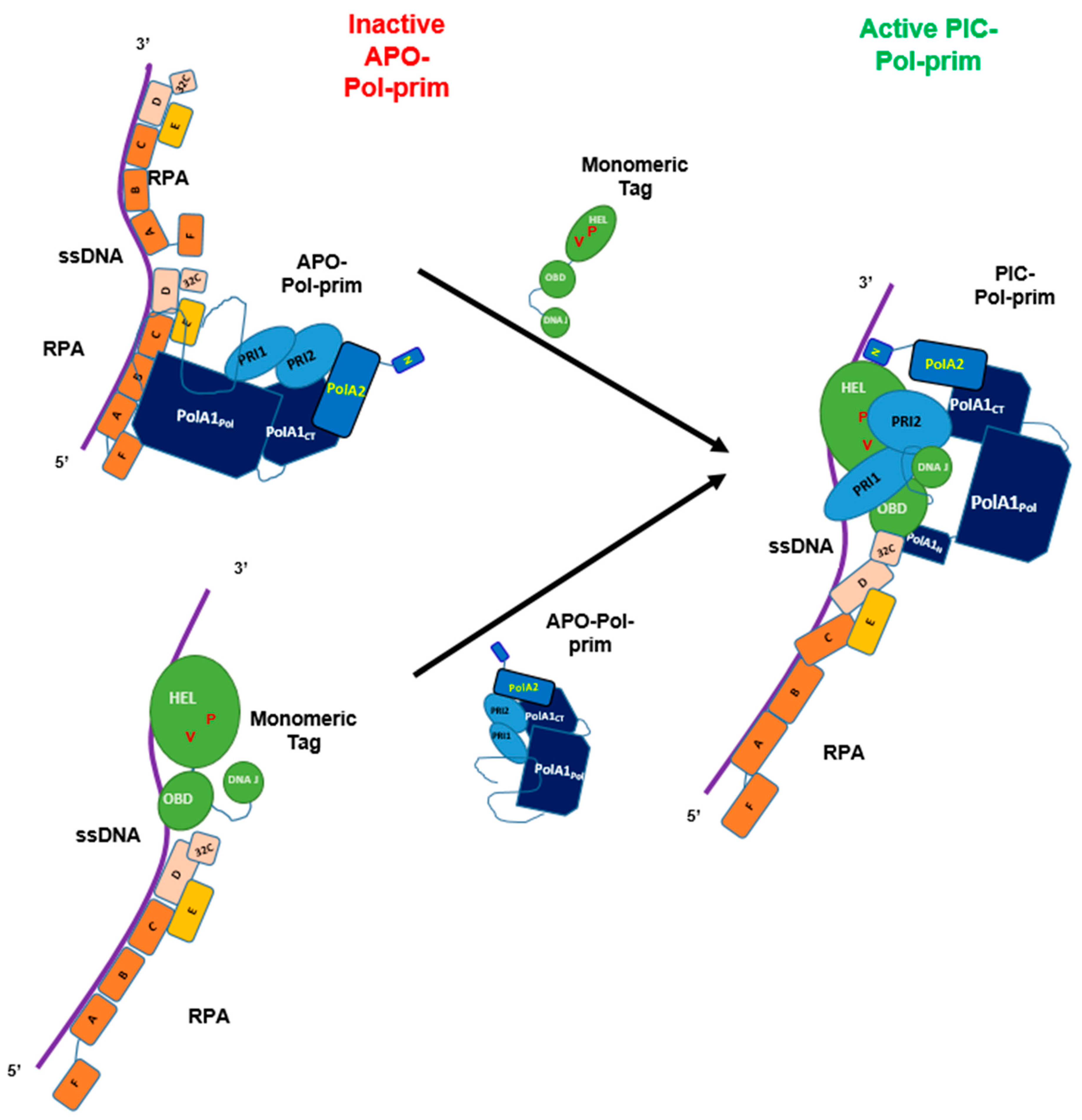
3.2. Lagging Strand Synthesis and the Initiation of Okazaki Fragment Synthesis
4. Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bleichert, F.; Botchan, M.R.; Berger, J.M. Mechanisms for initiating cellular DNA replication. Science 2017, 355, eaah6317. [Google Scholar] [CrossRef]
- Burgers, P.M.J.; Kunkel, T.A. Eukaryotic DNA Replication Fork. Annu. Rev. Biochem. 2017, 86, 417–438. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; O’Donnell, M. The Eukaryotic Replication Machine. Enzymes 2016, 39, 191–229. [Google Scholar] [CrossRef] [PubMed]
- Blow, J.J.; Laskey, R.A. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature 1988, 332, 546–548. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.J.; Cech, T.R. Shaping human telomeres: From shelterin and CST complexes to telomeric chromatin organization. Nat. Rev. Mol. Cell Biol. 2021, 22, 283–298. [Google Scholar] [CrossRef]
- Costa, A.; Diffley, J.F.X. The Initiation of Eukaryotic DNA Replication. Annu. Rev. Biochem. 2022, 91, 107–131. [Google Scholar] [CrossRef]
- Hu, Y.; Stillman, B. Origins of DNA replication in eukaryotes. Mol. Cell 2023, 83, 352–372. [Google Scholar] [CrossRef]
- Anderson, S.; DePamphilis, M.L. Metabolism of Okazaki fragments during simian virus 40 DNA replication. J. Biol. Chem. 1979, 254, 11495–11504. [Google Scholar] [CrossRef]
- Smith, D.J.; Whitehouse, I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature 2012, 483, 434–438. [Google Scholar] [CrossRef]
- Nasheuer, H.P.; Smith, R.; Bauerschmidt, C.; Grosse, F.; Weisshart, K. Initiation of eukaryotic DNA replication: Regulation and mechanisms. Prog. Nucleic Acid Res. Mol. Biol. 2002, 72, 41–94. [Google Scholar]
- Guilliam, T.A. Mechanisms for Maintaining Eukaryotic Replisome Progression in the Presence of DNA Damage. Front. Mol. Biosci. 2021, 8, 712971. [Google Scholar] [CrossRef]
- Nasheuer, H.P.; Pospiech, H.; Syväoja, J. Progress towards the anatomy of the eukaryotic DNA replication fork. In Genome Integrity: Facets and Perspectives; Genome Dynamics & Stability; Lankenau, D.H., Ed.; Springer: Berlin/Heidelberg, Geramny; New York, NY, USA, 2007; Volume 1, pp. 27–68. [Google Scholar]
- Baranovskiy, A.G.; Tahirov, T.H. Elaborated Action of the Human Primosome. Genes 2017, 8, 62. [Google Scholar] [CrossRef] [PubMed]
- Baranovskiy, A.G.; Zhang, Y.; Suwa, Y.; Babayeva, N.D.; Gu, J.; Pavlov, Y.I.; Tahirov, T.H. Crystal structure of the human primase. J. Biol. Chem. 2015, 290, 5635–5646. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.W.; Zinder, J.C.; Svetlov, V.; Bush, M.W.; Nudler, E.; Walz, T.; de Lange, T. Cryo-EM structure of the human CST–Polα/primase complex in a recruitment state. Nat. Struct. Mol. Biol. 2022, 29, 813–819. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Lin, X.; Chavez, B.L.; Agrawal, S.; Lusk, B.L.; Lim, C.J. Structures of the human CST-Polα–primase complex bound to telomere templates. Nature 2022, 608, 826–832. [Google Scholar] [CrossRef]
- Stadlbauer, F.; Brueckner, A.; Rehfuess, C.; Eckerskorn, C.; Lottspeich, F.; Förster, V.; Tseng, B.Y.; Nasheuer, H.P. DNA replication in vitro by recombinant DNA-polymerase-α-primase. Eur. J. Biochem. 1994, 222, 781–793. [Google Scholar] [CrossRef]
- Núñez-Ramírez, R.; Klinge, S.; Sauguet, L.; Melero, R.; Recuero-Checa, M.A.; Kilkenny, M.; Perera, R.L.; García-Alvarez, B.; Hall, R.J.; Nogales, E.; et al. Flexible tethering of primase and DNA Pol α in the eukaryotic primosome. Nucleic Acids Res. 2011, 39, 8187–8199. [Google Scholar] [CrossRef]
- Wong, S.W.; Wahl, A.F.; Yuan, P.-M.; Arai, N.; Pearson, B.E.; Arai, K.-I.; Korn, D.; Hunkapillar, M.W.; Wang, T.S.-F. Human DNA polymerase a gene expression is cell proliferation dependent and its primary structure is similar to both prokaryotic and eukaryotic replicative DNA polymerases. EMBO J. 1988, 7, 37–47. [Google Scholar] [CrossRef]
- Dephoure, N.; Zhou, C.; Villén, J.; Beausoleil, S.A.; Bakalarski, C.E.; Elledge, S.J.; Gygi, S.P. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA 2008, 105, 10762–10767. [Google Scholar] [CrossRef]
- Schub, O.; Rohaly, G.; Smith, R.W.; Schneider, A.; Dehde, S.; Dornreiter, I.; Nasheuer, H.P. Multiple phosphorylation sites of DNA polymerase α-primase cooperate to regulate the initiation of DNA replication in vitro. J. Biol. Chem. 2001, 276, 38076–38083. [Google Scholar] [CrossRef]
- Kilkenny, M.L.; Simon, A.C.; Mainwaring, J.; Wirthensohn, D.; Holzer, S.; Pellegrini, L. The human CTF4-orthologue AND-1 interacts with DNA polymerase α/primase via its unique C-terminal HMG box. Open Biol. 2017, 7. [Google Scholar] [CrossRef]
- Collins, K.L.; Russo, A.A.R.; Tseng, B.Y.; Kelly, T.J. The role of the 70 kDa subunit of human DNA polymerase a in DNA replication. EMBO J. 1993, 12, 4555–4566. [Google Scholar] [CrossRef] [PubMed]
- Voitenleitner, C.; Rehfuess, C.; Hilmes, M.; O’Rear, L.; Liao, P.C.; Gage, D.A.; Ott, R.; Nasheuer, H.P.; Fanning, E. Cell Cycle-Dependent Regulation of Human DNA Polymerase α-Primase Activity by Phosphorylation. Mol. Cell. Biol. 1999, 19, 646–656. [Google Scholar] [CrossRef]
- Voitenleitner, C.; Fanning, E.; Nasheuer, H.P. Phosphorylation of DNA polymerase α-primase by cyclin A-dependent kinases regulates initiation of DNA replication in vitro. Oncogene 1997, 14, 1611–1615. [Google Scholar] [CrossRef]
- Zhou, B.; Arnett, D.R.; Yu, X.; Brewster, A.; Sowd, G.A.; Xie, C.L.; Vila, S.; Gai, D.; Fanning, E.; Chen, X.S. Structural basis for the interaction of a hexameric replicative helicase with the regulatory subunit of human DNA polymerase alpha-primase. J. Biol. Chem. 2012, 287, 26854–26866. [Google Scholar] [CrossRef]
- Nasheuer, H.P.; Moore, A.; Wahl, A.F.; Wang, T.S. Cell cycle-dependent phosphorylation of human DNA polymerase a. J. Biol. Chem. 1991, 266, 7893–7903. [Google Scholar] [CrossRef]
- Nasheuer, H.P.; Grosse, F. DNA polymerase a-primase from calf thymus. Determination of the polypeptide responsible for primase activity. J. Biol. Chem. 1988, 263, 8981–8988. [Google Scholar] [CrossRef]
- Schneider, A.; Smith, R.W.P.; Kautz, A.R.; Weisshart, K.; Grosse, F.; Nasheuer, H.P. Primase activity of human DNA polymerase α-primase. Divalent cations stabilize the enzyme activity of the p48 subunit. J. Biol. Chem. 1998, 273, 21608–21615. [Google Scholar] [CrossRef]
- Zerbe, L.K.; Kuchta, R.D. The p58 subunit of human DNA primase is important for primer initiation, elongation, and counting. Biochemistry 2002, 41, 4891–4900. [Google Scholar] [CrossRef] [PubMed]
- Donnianni, R.A.; Zhou, Z.X.; Lujan, S.A.; Al-Zain, A.; Garcia, V.; Glancy, E.; Burkholder, A.B.; Kunkel, T.A.; Symington, L.S. DNA Polymerase Delta Synthesizes Both Strands during Break-Induced Replication. Mol. Cell 2019, 76, 371–381.e374. [Google Scholar] [CrossRef] [PubMed]
- Baretić, D.; Jenkyn-Bedford, M.; Aria, V.; Cannone, G.; Skehel, M.; Yeeles, J.T.P. Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork. Mol. Cell 2020, 78, 926–940.e913. [Google Scholar] [CrossRef]
- Bauerschmidt, C.; Pollok, S.; Kremmer, E.; Nasheuer, H.P.; Grosse, F. Interactions of human Cdc45 with the Mcm2-7 complex, the GINS complex, and DNA polymerases delta and epsilon during S phase. Genes Cells 2007, 12, 745–758. [Google Scholar] [PubMed]
- Gambus, A.; Jones, R.C.; Sanchez-Diaz, A.; Kanemaki, M.; van Deursen, F.; Edmondson, R.D.; Labib, K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 2006, 8, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Georgescu, R.E.; Langston, L.; Yao, N.Y.; Yurieva, O.; Zhang, D.; Finkelstein, J.; Agarwal, T.; O’Donnell, M.E. Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork. Nat. Struct. Mol. Biol. 2014, 21, 664–670. [Google Scholar] [CrossRef]
- Baris, Y.; Taylor, M.R.G.; Aria, V.; Yeeles, J.T.P. Fast and efficient DNA replication with purified human proteins. Nature 2022, 606, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Ilves, I.; Petojevic, T.; Pesavento, J.J.; Botchan, M.R. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol. Cell 2010, 37, 247–258. [Google Scholar] [CrossRef]
- Gambus, A.; van Deursen, F.; Polychronopoulos, D.; Foltman, M.; Jones, R.C.; Edmondson, R.D.; Calzada, A.; Labib, K. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J. 2009, 28, 2992–3004. [Google Scholar] [CrossRef]
- Broderick, R.; Nasheuer, H.P. Regulation of Cdc45 in the cell cycle and after DNA damage. Biochem. Soc. Trans. 2009, 37, 926–930. [Google Scholar] [CrossRef]
- Dornreiter, I.; Copeland, W.C.; Wang, T.S. Initiation of simian virus 40 DNA replication requires the interaction of a specific domain of human DNA polymerase alpha with large T antigen. Mol. Cell. Biol. 1993, 13, 809–820. [Google Scholar] [CrossRef]
- Villa, F.; Simon, A.C.; Ortiz Bazan, M.A.; Kilkenny, M.L.; Wirthensohn, D.; Wightman, M.; Matak-Vinkovíc, D.; Pellegrini, L.; Labib, K. Ctf4 Is a Hub in the Eukaryotic Replisome that Links Multiple CIP-Box Proteins to the CMG Helicase. Mol. Cell 2016, 63, 385–396. [Google Scholar] [CrossRef]
- Ning, B.; Feldkamp, M.D.; Cortez, D.; Chazin, W.J.; Friedman, K.L.; Fanning, E. Simian virus Large T antigen interacts with the N-terminal domain of the 70 kD subunit of Replication Protein A in the same mode as multiple DNA damage response factors. PLoS ONE 2015, 10, e0116093. [Google Scholar] [CrossRef] [PubMed]
- Robertson, P.D.; Warren, E.M.; Zhang, H.; Friedman, D.B.; Lary, J.W.; Cole, J.L.; Tutter, A.V.; Walter, J.C.; Fanning, E.; Eichman, B.F. Domain architecture and biochemical characterization of vertebrate Mcm10. J. Biol. Chem. 2008, 283, 3338–3348. [Google Scholar] [CrossRef]
- Ricke, R.M.; Bielinsky, A.K. Mcm10 regulates the stability and chromatin association of DNA polymerase-alpha. Mol. Cell 2004, 16, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Fien, K.; Cho, Y.S.; Lee, J.K.; Raychaudhuri, S.; Tappin, I.; Hurwitz, J. Primer utilization by DNA polymerase alpha-primase is influenced by its interaction with Mcm10p. J. Biol. Chem. 2004, 279, 16144–16153. [Google Scholar] [CrossRef]
- Broderick, R.; Rainey, M.D.; Santocanale, C.; Nasheuer, H.P. Cell cycle-dependent formation of Cdc45-Claspin complexes in human cells are compromized by UV-mediated DNA damage. FEBS J. 2013, 280, 4888–4902. [Google Scholar] [CrossRef]
- Broderick, S.; Rehmet, K.; Concannon, C.; Nasheuer, H.P. Eukaryotic single-stranded DNA binding proteins: Central factors in genome stability. Subcell. Biochem. 2010, 50, 143–163. [Google Scholar] [PubMed]
- Chen, R.; Wold, M.S. Replication protein A: Single-stranded DNA’s first responder: Dynamic DNA-interactions allow replication protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair. Bioessays 2014, 36, 1156–1161. [Google Scholar] [CrossRef]
- Wold, M.S. Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997, 66, 61–92. [Google Scholar] [CrossRef]
- Marechal, A.; Zou, L. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 2015, 25, 9–23. [Google Scholar] [CrossRef]
- Weisshart, K.; Pestryakov, P.; Smith, R.W.; Hartmann, H.; Kremmer, E.; Lavrik, O.; Nasheuer, H.P. Coordinated regulation of replication protein A activities by its subunits p14 and p32. J. Biol. Chem. 2004, 279, 35368–35376. [Google Scholar] [CrossRef]
- Williams, J.S.; Tumbale, P.P.; Arana, M.E.; Rana, J.A.; Williams, R.S.; Kunkel, T.A. High-fidelity DNA ligation enforces accurate Okazaki fragment maturation during DNA replication. Nat. Commun. 2021, 12, 482. [Google Scholar] [CrossRef]
- Casteel, D.E.; Zhuang, S.; Zeng, Y.; Perrino, F.W.; Boss, G.R.; Goulian, M.; Pilz, R.B. A DNA Polymerase-α·Primase Cofactor with Homology to Replication Protein A-32 Regulates DNA Replication in Mammalian Cells. J. Biol. Chem. 2009, 284, 5807–5818. [Google Scholar] [CrossRef]
- He, Y.; Song, H.; Chan, H.; Liu, B.; Wang, Y.; Sušac, L.; Zhou, Z.H.; Feigon, J. Structure of Tetrahymena telomerase-bound CST with polymerase α-primase. Nature 2022, 608, 813–818. [Google Scholar] [CrossRef]
- Huang, H.; Weiner, B.E.; Zhang, H.; Fuller, B.E.; Gao, Y.; Wile, B.M.; Zhao, K.; Arnett, D.R.; Chazin, W.J.; Fanning, E. Structure of a DNA polymerase alpha-primase domain that docks on the SV40 helicase and activates the viral primosome. J. Biol. Chem. 2010, 285, 17112–17122. [Google Scholar] [CrossRef]
- Olson, C.L.; Barbour, A.T.; Wuttke, D.S. Filling in the blanks: How the C-strand catches up to the G-strand at replicating telomeres using CST. Nat. Struct. Mol. Biol. 2022, 29, 730–733. [Google Scholar] [CrossRef]
- Onwubiko, N.O.; Borst, A.; Diaz, S.A.; Passkowski, K.; Scheffel, F.; Tessmer, I.; Nasheuer, H.P. SV40 T antigen interactions with ssDNA and replication protein A: A regulatory role of T antigen monomers in lagging strand DNA replication. Nucleic Acids Res. 2020, 48, 3657–3677. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.S.; Spenkelink, L.M.; Schauer, G.D.; Yurieva, O.; Mueller, S.H.; Natarajan, V.; Kaur, G.; Maher, C.; Kay, C.; O’Donnell, M.E.; et al. Tunability of DNA Polymerase Stability during Eukaryotic DNA Replication. Mol. Cell 2020, 77, 17–25.e15. [Google Scholar] [CrossRef]
- Onwubiko, N.O.; Scheffel, F.; Tessmer, I.; Nasheuer, H.P. SV40 T antigen helicase domain regions responsible for oligomerisation regulate Okazaki fragment synthesis initiation. FEBS Open Bio 2022, 12, 649–663. [Google Scholar] [CrossRef]
- Sun, J.; Yuan, Z.; Georgescu, R.; Li, H.; O’Donnell, M. The eukaryotic CMG helicase pumpjack and integration into the replisome. Nucleus 2016, 7, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Vaithiyalingam, S.; Arnett, D.R.; Aggarwal, A.; Eichman, B.F.; Fanning, E.; Chazin, W.J. Insights into eukaryotic primer synthesis from structures of the p48 subunit of human DNA primase. J. Mol. Biol. 2014, 426, 558–569. [Google Scholar] [CrossRef] [PubMed]
- Yeeles, J.T.P.; Janska, A.; Early, A.; Diffley, J.F.X. How the Eukaryotic Replisome Achieves Rapid and Efficient DNA Replication. Mol. Cell 2017, 65, 105–116. [Google Scholar] [CrossRef]
- Zaug, A.J.; Goodrich, K.J.; Song, J.J.; Sullivan, A.E.; Cech, T.R. Reconstitution of a telomeric replicon organized by CST. Nature 2022, 608, 819–825. [Google Scholar] [CrossRef]
- Lim, C.J.; Barbour, A.T.; Zaug, A.J.; Goodrich, K.J.; McKay, A.E.; Wuttke, D.S.; Cech, T.R. The structure of human CST reveals a decameric assembly bound to telomeric DNA. Science 2020, 368, 1081–1085. [Google Scholar] [CrossRef] [PubMed]
- de Lange, T. Shelterin-Mediated Telomere Protection. Annu. Rev. Genet. 2018, 52, 223–247. [Google Scholar] [CrossRef]
- Diede, S.J.; Gottschling, D.E. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 1999, 99, 723–733. [Google Scholar] [CrossRef]
- Lue, N.F.; Chan, J.; Wright, W.E.; Hurwitz, J. The CDC13-STN1-TEN1 complex stimulates Pol alpha activity by promoting RNA priming and primase-to-polymerase switch. Nat. Commun. 2014, 5, 5762. [Google Scholar] [CrossRef]
- Nakaoka, H.; Nishiyama, A.; Saito, M.; Ishikawa, F. Xenopus laevis Ctc1-Stn1-Ten1 (xCST) protein complex is involved in priming DNA synthesis on single-stranded DNA template in Xenopus egg extract. J. Biol. Chem. 2012, 287, 619–627. [Google Scholar] [CrossRef]
- Baranovskiy, A.G.; Babayeva, N.D.; Zhang, Y.; Gu, J.; Suwa, Y.; Pavlov, Y.I.; Tahirov, T.H. Mechanism of Concerted RNA-DNA Primer Synthesis by the Human Primosome. J. Biol. Chem. 2016, 291, 10006–10020. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Ganduri, S.; Lue, N.F. STN1-POLA2 interaction provides a basis for primase-pol α stimulation by human STN1. Nucleic Acids Res. 2017, 45, 9455–9466. [Google Scholar] [CrossRef]
- Stewart, J.A.; Wang, F.; Chaiken, M.F.; Kasbek, C.; Chastain, P.D., 2nd; Wright, W.E.; Price, C.M. Human CST promotes telomere duplex replication and general replication restart after fork stalling. EMBO J. 2012, 31, 3537–3549. [Google Scholar] [CrossRef] [PubMed]
- Kasbek, C.; Wang, F.; Price, C.M. Human TEN1 maintains telomere integrity and functions in genome-wide replication restart. J. Biol. Chem. 2013, 288, 30139–30150. [Google Scholar] [CrossRef]
- Nieminuszczy, J.; Broderick, R.; Niedzwiedz, W. EXD2—A new player joins the DSB resection team. Cell Cycle 2016, 15, 1519–1520. [Google Scholar] [CrossRef] [PubMed]
- Mirman, Z.; Cai, S.; de Lange, T. CST/Polα/primase-mediated fill-in synthesis at DSBs. Cell Cycle 2023, 22, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Mirman, Z.; Lottersberger, F.; Takai, H.; Kibe, T.; Gong, Y.; Takai, K.; Bianchi, A.; Zimmermann, M.; Durocher, D.; de Lange, T. 53BP1-RIF1-shieldin counteracts DSB resection through CST- and Polα-dependent fill-in. Nature 2018, 560, 112–116. [Google Scholar] [CrossRef]
- Mirman, Z.; Sasi, N.K.; King, A.; Chapman, J.R.; de Lange, T. 53BP1-shieldin-dependent DSB processing in BRCA1-deficient cells requires CST-Polα-primase fill-in synthesis. Nat. Cell Biol. 2022, 24, 51–61. [Google Scholar] [CrossRef]
- Noordermeer, S.M.; Adam, S.; Setiaputra, D.; Barazas, M.; Pettitt, S.J.; Ling, A.K.; Olivieri, M.; Álvarez-Quilón, A.; Moatti, N.; Zimmermann, M.; et al. The shieldin complex mediates 53BP1-dependent DNA repair. Nature 2018, 560, 117–121. [Google Scholar] [CrossRef]
- Miyake, Y.; Nakamura, M.; Nabetani, A.; Shimamura, S.; Tamura, M.; Yonehara, S.; Saito, M.; Ishikawa, F. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol. Cell 2009, 36, 193–206. [Google Scholar] [CrossRef]
- Arunkumar, A.I.; Klimovich, V.; Jiang, X.; Ott, R.D.; Mizoue, L.; Fanning, E.; Chazin, W.J. Insights into hRPA32 C-terminal domain--mediated assembly of the simian virus 40 replisome. Nat. Struct. Mol. Biol. 2005, 12, 332–339. [Google Scholar] [CrossRef]
- Weisshart, K.; Förster, H.; Kremmer, E.; Schlott, B.; Grosse, F.; Nasheuer, H.P. Protein-protein interactions of the primase subunits p58 and p48 with simian virus 40 T antigen are required for efficient primer synthesis in a cell-free system. J. Biol. Chem. 2000, 275, 17328–17337. [Google Scholar] [CrossRef]
- Melendy, T.; Stillman, B. An interaction between replication protein A and SV40 T antigen appears essential for primosome assembly during SV40 DNA replication. J. Biol. Chem. 1993, 268, 3389–3395. [Google Scholar] [CrossRef] [PubMed]
- Collins, K.L.; Kelly, T.J. The effects of T antigen and replication protein A on the initiation of DNA synthesis by DNA polymerase a-primase. Mol. Cell. Biol. 1991, 11, 2108–2115. [Google Scholar] [CrossRef] [PubMed]
- Challberg, M.; Kelly, T.J. Animal virus DNA replication. Annu. Rev. Biochem. 1989, 58, 671–717. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Kelly, T.J. Simian virus 40 DNA replication in vitro. Proc. Natl. Acad. Sci. USA 1984, 81, 6973–6977. [Google Scholar] [CrossRef] [PubMed]
- Sowd, G.A.; Fanning, E. A wolf in sheep’s clothing: SV40 co-opts host genome maintenance proteins to replicate viral DNA. PLoS Pathog. 2012, 8, e1002994. [Google Scholar] [CrossRef] [PubMed]
- Waga, S.; Bauer, G.; Stillman, B. Reconstitution of complete SV40 DNA replication with purified replication factors. J. Biol. Chem. 1994, 269, 10923–10934. [Google Scholar] [CrossRef]
- Waga, S.; Stillman, B. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 1998, 67, 721–751. [Google Scholar] [CrossRef]
- Jiang, X.; Klimovich, V.; Arunkumar, A.I.; Hysinger, E.B.; Wang, Y.; Ott, R.D.; Guler, G.D.; Weiner, B.; Chazin, W.J.; Fanning, E. Structural mechanism of RPA loading on DNA during activation of a simple pre-replication complex. EMBO J. 2006, 25, 5516–5526. [Google Scholar] [CrossRef]
- Fanning, E.; Klimovich, V.; Nager, A.R. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006, 34, 4126–4137. [Google Scholar] [CrossRef]
- Fanning, E.; Knippers, R. Structure and function of simian virus 40 large tumor antigen. Annu. Rev. Biochem. 1992, 61, 55–85. [Google Scholar] [CrossRef] [PubMed]
- Braun, K.A.; Lao, Y.; He, Z.; Ingles, C.J.; Wold, M.S. Role of protein-protein interactions in the function of replication protein A (RPA): RPA modulates the activity of DNA polymerase a by multiple mechanisms. Biochemistry 1997, 36, 8443–8454. [Google Scholar] [CrossRef]
- Maga, G.; Frouin, I.; Spadari, S.; Hubscher, U. Replication protein A as a “fidelity clamp” for DNA polymerase alpha. J. Biol. Chem. 2001, 276, 18235–18242. [Google Scholar] [CrossRef] [PubMed]
- An, P.; Saenz Robles, M.T.; Pipas, J.M. Large T antigens of polyomaviruses: Amazing molecular machines. Annu. Rev. Microbiol. 2012, 66, 213–236. [Google Scholar] [CrossRef]
- Smith, R.W.; Steffen, C.; Grosse, F.; Nasheuer, H.P. Species specificity of simian virus 40 DNA replication in vitro requires multiple functions of human DNA polymerase alpha. J. Biol. Chem. 2002, 277, 20541–20548. [Google Scholar] [CrossRef]
- Stadlbauer, F.; Voitenleitner, C.; Brückner, A.; Fanning, E.; Nasheuer, H.P. Species-specific replication of simian virus 40 DNA in vitro requires the p180 subunit of human DNA polymerase a-primase. Mol. Cell. Biol. 1996, 16, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, T.; Hirabayashi, K.; Miyazawa, S.; Kobayashi, Y.; Shoji, K.; Kobayashi, M.; Hanaoka, F.; Imamoto, N.; Torigoe, H. The intrinsically disordered N-terminal region of mouse DNA polymerase alpha mediates its interaction with POT1a/b at telomeres. Genes Cells 2021, 26, 360–380. [Google Scholar] [CrossRef]
- Georgescu, R.E.; Schauer, G.D.; Yao, N.Y.; Langston, L.D.; Yurieva, O.; Zhang, D.; Finkelstein, J.; O’Donnell, M.E. Reconstitution of a eukaryotic replisome reveals suppression mechanisms that define leading/lagging strand operation. eLife 2015, 4, e04988. [Google Scholar] [CrossRef]
- Devbhandari, S.; Jiang, J.; Kumar, C.; Whitehouse, I.; Remus, D. Chromatin Constrains the Initiation and Elongation of DNA Replication. Mol. Cell 2017, 65, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Kurat, C.F.; Yeeles, J.T.P.; Patel, H.; Early, A.; Diffley, J.F.X. Chromatin Controls DNA Replication Origin Selection, Lagging-Strand Synthesis, and Replication Fork Rates. Mol. Cell 2017, 65, 117–130. [Google Scholar] [CrossRef]
- Yeeles, J.T.; Deegan, T.D.; Janska, A.; Early, A.; Diffley, J.F. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 2015, 519, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.L.; Baris, Y.; Taylor, M.R.G.; Yeeles, J.T.P. Structure of a human replisome shows the organisation and interactions of a DNA replication machine. EMBO J. 2021, 40, e108819. [Google Scholar] [CrossRef] [PubMed]
- Guilliam, T.A.; Yeeles, J.T.P. An updated perspective on the polymerase division of labor during eukaryotic DNA replication. Crit. Rev. Biochem. Mol. Biol. 2020, 55, 469–481. [Google Scholar] [CrossRef]
- Taylor, M.R.G.; Yeeles, J.T.P. Dynamics of Replication Fork Progression Following Helicase-Polymerase Uncoupling in Eukaryotes. J. Mol. Biol. 2019, 431, 2040–2049. [Google Scholar] [CrossRef]
- Szambowska, A.; Tessmer, I.; Prus, P.; Schlott, B.; Pospiech, H.; Grosse, F. Cdc45-induced loading of human RPA onto single-stranded DNA. Nucleic Acids Res. 2017, 45, 3217–3230. [Google Scholar] [CrossRef] [PubMed]
- Ho, B.; Baryshnikova, A.; Brown, G.W. Unification of Protein Abundance Datasets Yields a Quantitative Saccharomyces cerevisiae Proteome. Cell Syst. 2018, 6, 192–205.e193. [Google Scholar] [CrossRef]
- Zhu, W.; Ukomadu, C.; Jha, S.; Senga, T.; Dhar, S.K.; Wohlschlegel, J.A.; Nutt, L.K.; Kornbluth, S.; Dutta, A. Mcm10 and And-1/CTF4 recruit DNA polymerase alpha to chromatin for initiation of DNA replication. Genes Dev. 2007, 21, 2288–2299. [Google Scholar] [CrossRef] [PubMed]
- Goulian, M.; Heard, C.J. The mechanism of action of an accessory protein for DNA polymerase alpha/primase. J. Biol. Chem. 1990, 265, 13231–13239. [Google Scholar] [CrossRef]
- Surovtseva, Y.V.; Churikov, D.; Boltz, K.A.; Song, X.; Lamb, J.C.; Warrington, R.; Leehy, K.; Heacock, M.; Price, C.M.; Shippen, D.E. Conserved Telomere Maintenance Component 1 Interacts with STN1 and Maintains Chromosome Ends in Higher Eukaryotes. Mol. Cell 2009, 36, 207–218. [Google Scholar] [CrossRef]
- Wan, M.; Qin, J.; Songyang, Z.; Liu, D. OB fold-containing protein 1 (OBFC1), a human homolog of yeast Stn1, associates with TPP1 and is implicated in telomere length regulation. J. Biol. Chem. 2009, 284, 26725–26731. [Google Scholar] [CrossRef]
- Chandra, A.; Hughes, T.R.; Nugent, C.I.; Lundblad, V. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 2001, 15, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Kesti, T.; Flick, K.; Keränen, S.; Syväoja, J.E.; Wittenberg, C. DNA polymerase e catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol. Cell 1999, 3, 679–685. [Google Scholar] [CrossRef]
- DiFrancesco, R.A.; Lehman, I.R. Interaction of ribonuclease H from Drosophila melanogaster embryos with DNA polymerase-primase. J. Biol. Chem. 1985, 260, 14764–14770. [Google Scholar] [CrossRef] [PubMed]
- Frank, P.; Braunshofer-Reiter, C.; Karwan, A.; Grimm, R.; Wintersberger, U. Purification of Saccharomyces cerevisiae RNase H(70) and identification of the corresponding gene. FEBS Lett. 1999, 450, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Karwan, R.; Blutsch, H.; Wintersberger, U. A ribonuclease H from yeast stimulates DNA polymerase in vitro. Adv. Exp. Med. Biol. 1984, 179, 513–518. [Google Scholar] [CrossRef]
- Hagemeier, A.; Grosse, F. A distinct form of ribonuclease H from calf thymus stimulates its homologous DNA-polymerase-alpha-primase complex. Eur. J. Biochem. 1989, 185, 621–628. [Google Scholar] [CrossRef]
- De Falco, M.; Ferrari, E.; De Felice, M.; Rossi, M.; Hubscher, U.; Pisani, F.M. The human GINS complex binds to and specifically stimulates human DNA polymerase alpha-primase. EMBO Rep. 2007, 8, 99–103. [Google Scholar] [CrossRef]
- You, Z.; De Falco, M.; Kamada, K.; Pisani, F.; Masai, H. The mini-chromosome maintenance (Mcm) complexes interact with DNA polymerase alpha-primase and stimulate its ability to synthesize RNA primers. PLoS ONE 2013, 8, e72408. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nasheuer, H.P.; Onwubiko, N.O. Lagging Strand Initiation Processes in DNA Replication of Eukaryotes—Strings of Highly Coordinated Reactions Governed by Multiprotein Complexes. Genes 2023, 14, 1012. https://doi.org/10.3390/genes14051012
Nasheuer HP, Onwubiko NO. Lagging Strand Initiation Processes in DNA Replication of Eukaryotes—Strings of Highly Coordinated Reactions Governed by Multiprotein Complexes. Genes. 2023; 14(5):1012. https://doi.org/10.3390/genes14051012
Chicago/Turabian StyleNasheuer, Heinz Peter, and Nichodemus O. Onwubiko. 2023. "Lagging Strand Initiation Processes in DNA Replication of Eukaryotes—Strings of Highly Coordinated Reactions Governed by Multiprotein Complexes" Genes 14, no. 5: 1012. https://doi.org/10.3390/genes14051012





